![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 1530_Read1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 55605619 |
| Filtered Sequences | 0 |
| Sequence length | 75 |
| %GC | 42 |
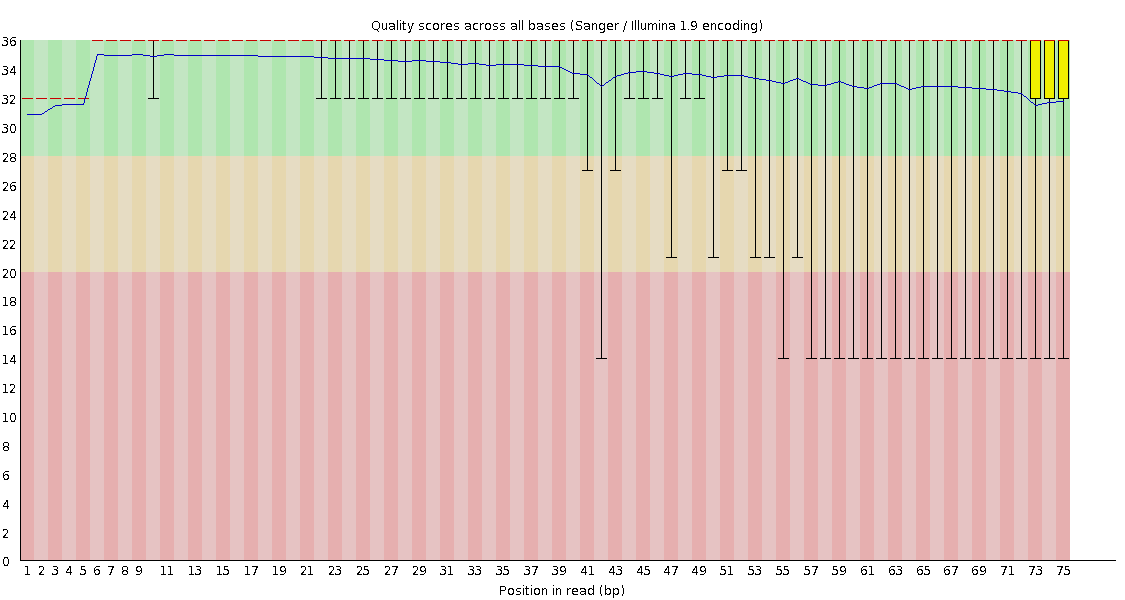
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
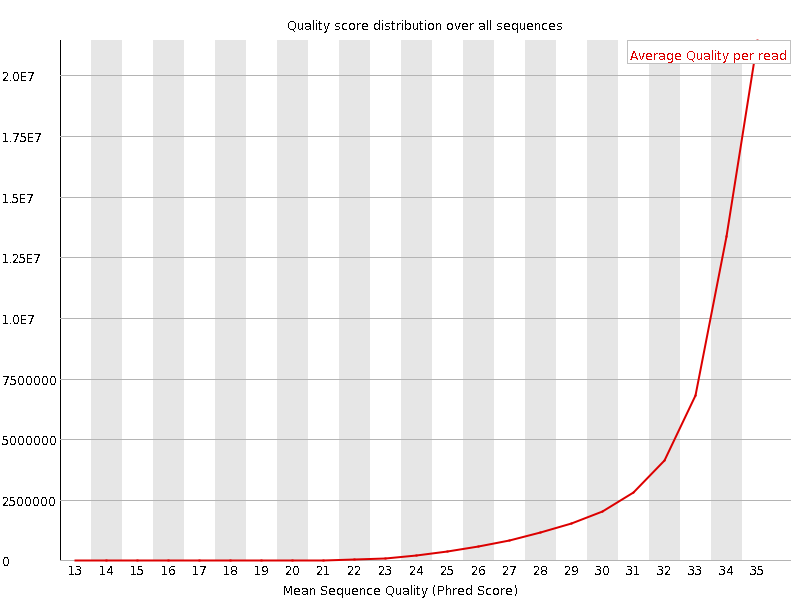
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

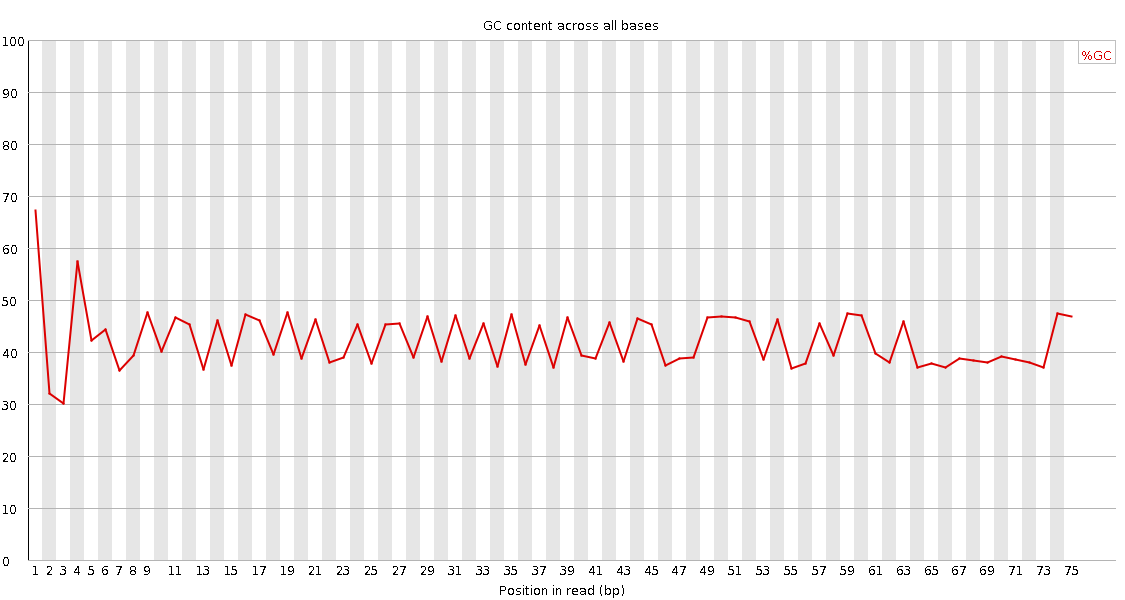
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
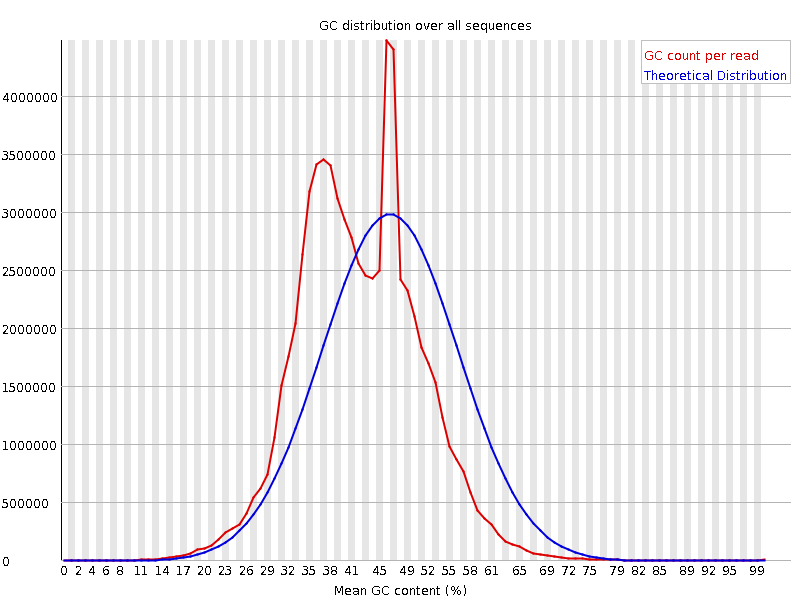
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
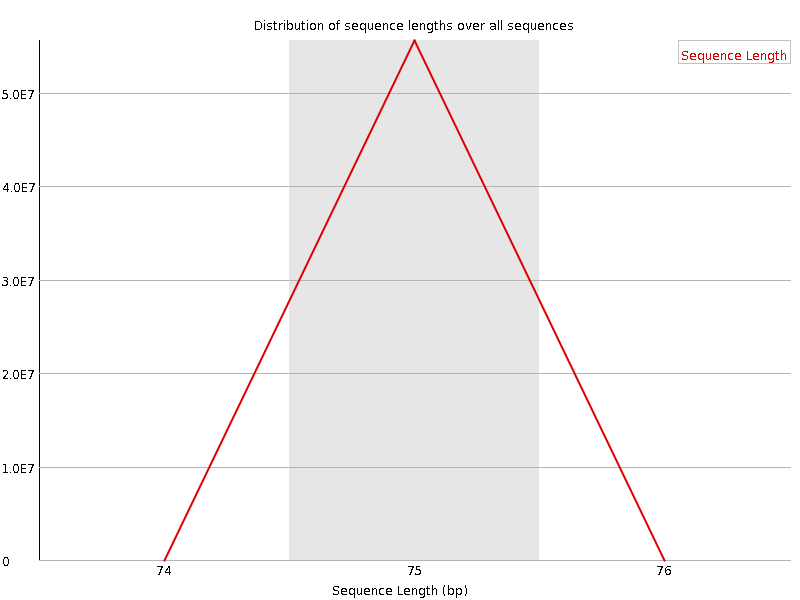
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
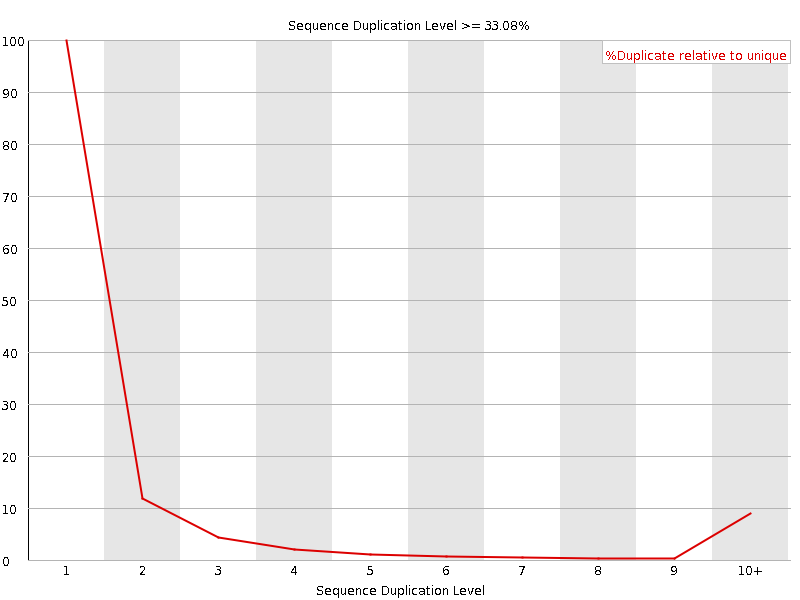
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACACAGTGATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAGG | 3734780 | 6.7165514334081955 | TruSeq Adapter, Index 5 (100% over 63bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACACAGTGATGTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAGG | 79548 | 0.1430574848919495 | TruSeq Adapter, Index 5 (98% over 63bp) |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
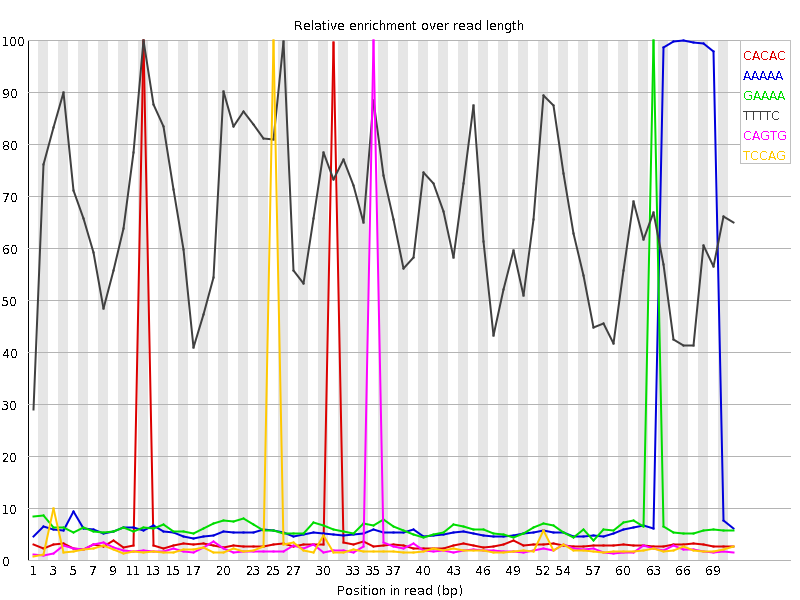
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CACAC | 18865800 | 5.7545877 | 100.56842 | 12 |
| AAAAA | 46593490 | 5.505951 | 40.781437 | 66 |
| GAAAA | 25453040 | 4.186632 | 55.975525 | 63 |
| TTTTC | 21811750 | 3.985155 | 6.004756 | 12 |
| CAGTG | 11635605 | 3.7653208 | 105.60204 | 35 |
| TCCAG | 11776345 | 3.75563 | 104.630554 | 25 |
| GTGTG | 10442240 | 3.5329707 | 8.74508 | 21 |
| CTGAA | 14604405 | 3.3953056 | 77.22915 | 19 |
| ACACA | 14849975 | 3.3020787 | 73.3625 | 32 |
| CGTCT | 9975880 | 3.2780538 | 106.013985 | 16 |
| TTTCT | 17686440 | 3.231433 | 6.234968 | 27 |
| CTCCA | 10074300 | 3.166259 | 102.40951 | 24 |
| TGAAA | 18458780 | 3.1283884 | 57.92399 | 62 |
| GGAAG | 9669245 | 3.08144 | 103.925575 | 5 |
| CACAG | 9955055 | 3.0812247 | 99.975006 | 33 |
| GATCG | 9044335 | 2.9267771 | 110.856544 | 1 |
| GCACA | 9370590 | 2.9003248 | 101.03306 | 11 |
| CACGT | 9072895 | 2.8934646 | 104.25522 | 14 |
| TCTGC | 8663225 | 2.8467183 | 106.568344 | 56 |
| GATCT | 11726180 | 2.8089507 | 73.28633 | 1 |
| CTGCT | 8509580 | 2.7962306 | 103.44561 | 57 |
| TGTGT | 11129590 | 2.787405 | 5.3451552 | 36 |
| TTCTG | 11278930 | 2.7838643 | 78.49009 | 55 |
| GTGGA | 8421290 | 2.765237 | 7.074621 | 39 |
| CCTGG | 6194945 | 2.7499688 | 7.283201 | 18 |
| CTTCT | 11212270 | 2.7273004 | 77.29322 | 54 |
| AGAGC | 8588470 | 2.6973436 | 102.64917 | 8 |
| GAGCA | 8542215 | 2.6828165 | 102.45539 | 9 |
| GTCTG | 8040045 | 2.680798 | 108.39329 | 17 |
| CCAGT | 8319455 | 2.6531827 | 102.97824 | 26 |
| TCTTC | 10786665 | 2.623775 | 77.30706 | 53 |
| TTTCC | 10737225 | 2.6117492 | 5.1211567 | 50 |
| TCCAC | 8284725 | 2.6038125 | 10.9562025 | 3 |
| TTCTC | 10672395 | 2.5959797 | 5.3555274 | 29 |
| TTCCA | 10941845 | 2.5830772 | 5.489504 | 51 |
| AGCAC | 8127655 | 2.5156198 | 100.861275 | 10 |
| GAAGA | 10899430 | 2.4954388 | 75.10941 | 6 |
| CAGTC | 7761095 | 2.4751146 | 102.91031 | 27 |
| TCTGA | 10265395 | 2.4590268 | 78.16794 | 18 |
| CTTGA | 10123095 | 2.4249396 | 76.13099 | 60 |
| ACGTC | 7591245 | 2.420947 | 103.87765 | 15 |
| ACTCC | 7698200 | 2.419473 | 101.73191 | 23 |
| GAGAA | 10494080 | 2.4026334 | 5.0060115 | 29 |
| ATGCC | 7404345 | 2.3613424 | 101.70562 | 47 |
| AAGAG | 10293400 | 2.3566873 | 75.20634 | 7 |
| AAAGG | 10286675 | 2.3551476 | 71.424644 | 71 |
| GTGAT | 9530925 | 2.3166656 | 77.02978 | 37 |
| GCTTG | 6874645 | 2.292218 | 105.15709 | 59 |
| TCTCT | 9349625 | 2.274226 | 7.7074714 | 3 |
| AGTGA | 9628385 | 2.2713766 | 75.620834 | 36 |
| GTCCT | 6903410 | 2.2684467 | 5.2084527 | 13 |
| TGCCG | 5052680 | 2.2429116 | 140.52505 | 48 |
| ACAGT | 9637060 | 2.2404723 | 75.0394 | 34 |
| TCACA | 9774630 | 2.2395182 | 74.45428 | 30 |
| GCCAT | 6962350 | 2.2203844 | 5.278625 | 1 |
| AGGAC | 7041965 | 2.2116394 | 6.806278 | 66 |
| CGGAA | 6990500 | 2.195476 | 101.84113 | 4 |
| CTGGG | 4871710 | 2.1943834 | 7.485921 | 19 |
| CCGTC | 5012935 | 2.1930153 | 138.00107 | 50 |
| GCCGT | 4907160 | 2.1783142 | 140.2156 | 49 |
| GTCAC | 6815890 | 2.1736765 | 102.74654 | 29 |
| TCTCA | 9178525 | 2.1668046 | 6.5519423 | 3 |
| ATGGC | 6576990 | 2.1283362 | 5.864689 | 12 |
| GGTGG | 4645620 | 2.12332 | 6.3111186 | 69 |
| CCTCC | 4888775 | 2.1077003 | 8.443049 | 4 |
| CTCTG | 6386550 | 2.0986073 | 5.345491 | 61 |
| CTCTC | 6454660 | 2.0902467 | 5.7774653 | 14 |
| CCCTG | 4737605 | 2.0725665 | 7.902842 | 4 |
| AGTGG | 6310190 | 2.0720305 | 6.20207 | 57 |
| ATCTC | 8754955 | 2.0668108 | 71.72055 | 40 |
| TCCTC | 6353270 | 2.0574129 | 10.698348 | 3 |
| TCTCG | 6255645 | 2.0555923 | 100.10265 | 41 |
| GATCC | 6442550 | 2.0546134 | 98.32074 | 1 |
| CCCAG | 4834115 | 2.0524566 | 6.3659253 | 70 |
| TGCTT | 8303115 | 2.049374 | 77.8696 | 58 |
| GTCTT | 8216985 | 2.0281155 | 78.10907 | 52 |
| AACTC | 8745010 | 2.0036163 | 74.50183 | 22 |
| GAACT | 8527385 | 1.9824893 | 75.61419 | 21 |
| CAGAG | 6242035 | 1.9604089 | 5.803199 | 54 |
| AAAAG | 11867135 | 1.9519604 | 51.98618 | 70 |
| CCACC | 4643940 | 1.9431347 | 7.2807655 | 4 |
| CCACT | 6142090 | 1.930402 | 5.442654 | 29 |
| GAGGA | 5974880 | 1.9041026 | 5.794148 | 32 |
| CTGTG | 5701865 | 1.9011769 | 5.778701 | 19 |
| AGTCA | 8173755 | 1.9002757 | 75.2041 | 28 |
| CAGGG | 4333735 | 1.8945272 | 5.781024 | 6 |
| CTCCT | 5771055 | 1.8688712 | 5.5625224 | 16 |
| TCGGA | 5741080 | 1.8578328 | 104.73056 | 3 |
| ATCGG | 5672185 | 1.8355383 | 105.74539 | 2 |
| GCTGG | 4034360 | 1.8172125 | 7.482436 | 27 |
| ATCCA | 7883395 | 1.8062072 | 28.81138 | 2 |
| ACACG | 5789540 | 1.7919413 | 100.127075 | 13 |
| TGAAC | 7693120 | 1.7885352 | 75.36853 | 20 |
| TCCAT | 7557680 | 1.7841663 | 11.6874075 | 3 |
| TCCTT | 7331695 | 1.7833797 | 8.731483 | 3 |
| TTGAA | 10060260 | 1.7567885 | 56.170708 | 61 |
| CCTTC | 5392905 | 1.7464129 | 5.045684 | 4 |
| GGAGG | 3909410 | 1.734165 | 6.089992 | 19 |
| CCAGC | 4082610 | 1.7333845 | 6.987879 | 4 |
| CTGAG | 5294765 | 1.7134037 | 5.683926 | 30 |
| CCTCT | 5266675 | 1.7055352 | 5.450675 | 13 |
| GTATG | 6979980 | 1.6966119 | 77.464226 | 45 |
| TCCTG | 5160485 | 1.6957248 | 10.821854 | 3 |
| TGCTG | 5049260 | 1.6835784 | 5.947525 | 26 |
| CTGGC | 3779145 | 1.6775824 | 5.539883 | 28 |
| TCTGT | 6772895 | 1.6716852 | 6.400078 | 3 |
| CATCC | 5302190 | 1.6664292 | 8.916079 | 5 |
| GGCAG | 3796260 | 1.6595656 | 6.285997 | 14 |
| TCTGG | 4971870 | 1.6577742 | 6.2943497 | 3 |
| CCATA | 7216985 | 1.6535224 | 5.1117105 | 9 |
| CCAGA | 5320640 | 1.6468104 | 5.2746506 | 53 |
| TATGC | 6871265 | 1.645979 | 76.262054 | 46 |
| CTCGT | 4959240 | 1.6295961 | 100.35142 | 42 |
| TCCCA | 5105560 | 1.6046301 | 8.294684 | 3 |
| TCAAG | 6880185 | 1.59954 | 8.915595 | 3 |
| GTGGG | 3496990 | 1.598329 | 5.8295913 | 70 |
| GGACC | 3662090 | 1.5777086 | 7.8281145 | 67 |
| GCCAG | 3613305 | 1.5566908 | 8.153536 | 40 |
| TGTCC | 4725870 | 1.5529113 | 5.510891 | 19 |
| TCTTT | 8482025 | 1.5497236 | 9.2732725 | 3 |
| TGATC | 6461825 | 1.5478995 | 72.72381 | 38 |
| TCCCT | 4739360 | 1.5347719 | 11.694258 | 3 |
| TGGAG | 4670585 | 1.5336457 | 5.0994434 | 40 |
| TCTCC | 4714175 | 1.5266162 | 7.1455708 | 3 |
| AGGTG | 4642060 | 1.5242792 | 5.092598 | 13 |
| TCCCC | 3518395 | 1.5168877 | 8.190424 | 3 |
| GGTCC | 3371165 | 1.496478 | 6.998162 | 28 |
| GCTCC | 3413165 | 1.4931618 | 6.327361 | 47 |
| GCCTG | 3351175 | 1.4876043 | 5.1029425 | 70 |
| TCAGG | 4595825 | 1.4872246 | 5.74194 | 3 |
| AGCTG | 4485105 | 1.451395 | 5.032914 | 13 |
| GCCCC | 2441540 | 1.4219952 | 6.779448 | 48 |
| GATCA | 6096535 | 1.4173532 | 59.62321 | 1 |
| TCCTA | 5974945 | 1.4105248 | 5.091121 | 3 |
| TGGTG | 4122155 | 1.3946675 | 5.818895 | 19 |
| CCATC | 4400740 | 1.3831118 | 9.695733 | 4 |
| GTGAG | 4172420 | 1.370067 | 5.0433526 | 21 |
| GCAAG | 4327095 | 1.358992 | 6.4300485 | 50 |
| GGGCA | 3103295 | 1.3566304 | 6.4926085 | 21 |
| GGCCC | 2288985 | 1.3527516 | 7.780249 | 47 |
| GGAGC | 3091345 | 1.3514063 | 5.645413 | 22 |
| CACCA | 4416245 | 1.347076 | 5.8346167 | 33 |
| TGGGC | 2970810 | 1.3381535 | 6.5175834 | 20 |
| TGAGG | 4017275 | 1.319123 | 5.307834 | 31 |
| AGGCC | 3044535 | 1.3116524 | 5.9224706 | 46 |
| GCTGA | 3930025 | 1.2717693 | 5.364758 | 28 |
| GTCCA | 3974720 | 1.26759 | 5.120996 | 20 |
| CAAGG | 4006855 | 1.2584155 | 5.024609 | 4 |
| ATCAC | 5490625 | 1.2579868 | 11.921855 | 2 |
| ATCCC | 3949890 | 1.241414 | 24.245258 | 2 |
| ATCTG | 5155570 | 1.2349923 | 19.014454 | 2 |
| GTCTC | 3742200 | 1.2296795 | 7.9020324 | 59 |
| CGTAT | 5087655 | 1.2187237 | 75.86788 | 44 |
| ATCCT | 5082285 | 1.1997917 | 24.767813 | 2 |
| CCCAT | 3775630 | 1.1866456 | 6.5679193 | 8 |
| TCAGC | 3688520 | 1.1763173 | 5.9616847 | 15 |
| TCCAA | 5131700 | 1.1757514 | 7.014587 | 21 |
| TCGTA | 4899250 | 1.1735921 | 75.35488 | 43 |
| TCATG | 4805240 | 1.1510724 | 5.4030538 | 3 |
| AGGGC | 2607670 | 1.139964 | 5.2408457 | 7 |
| CTGAC | 3505120 | 1.1178285 | 5.452969 | 29 |
| ATCTT | 6167500 | 1.0936322 | 20.993387 | 2 |
| GTGGT | 3217860 | 1.0887132 | 5.125064 | 58 |
| ATCAG | 4595125 | 1.0682977 | 12.305138 | 2 |
| GACAC | 3415540 | 1.0571561 | 6.2637396 | 31 |
| ACACC | 3223040 | 0.9831158 | 5.4703746 | 32 |
| ATCAT | 5238355 | 0.90149736 | 11.288941 | 2 |
| TCAAA | 5174690 | 0.8642936 | 6.2457776 | 3 |
| GACCC | 2002120 | 0.8500552 | 6.8262267 | 68 |
| ATCAA | 5034315 | 0.84084773 | 16.261328 | 2 |
| CTAGC | 2468895 | 0.78736293 | 5.2446017 | 47 |
| TCTTA | 3921285 | 0.6953293 | 5.23234 | 3 |
| ATCTA | 3618775 | 0.6227749 | 8.103573 | 2 |
| CCCGG | 1018495 | 0.6019134 | 5.2998695 | 50 |
| CCCCG | 990770 | 0.5770417 | 5.274226 | 49 |
| CCGCC | 803820 | 0.4681587 | 5.4604783 | 38 |
| CCGGC | 662265 | 0.39138743 | 5.1306205 | 51 |