| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | KLF2 | Kruppel like factor 2 [Source:HGNC Symbol;Acc:HGNC:6347] | 30 | 7.567 | 0.0319 | No |
| 2 | CHST12 | carbohydrate sulfotransferase 12 [Source:HGNC Symbol;Acc:HGNC:17423] | 65 | 6.683 | 0.0562 | No |
| 3 | EDARADD | EDAR associated death domain [Source:HGNC Symbol;Acc:HGNC:14341] | 80 | 6.469 | 0.0894 | No |
| 4 | MMAA | metabolism of cobalamin associated A [Source:HGNC Symbol;Acc:HGNC:18871] | 313 | 4.488 | -0.0022 | No |
| 5 | HIC1 | HIC ZBTB transcriptional repressor 1 [Source:HGNC Symbol;Acc:HGNC:4909] | 459 | 3.975 | -0.0522 | No |
| 6 | RNF25 | ring finger protein 25 [Source:HGNC Symbol;Acc:HGNC:14662] | 592 | 3.666 | -0.0973 | No |
| 7 | FBXO32 | F-box protein 32 [Source:HGNC Symbol;Acc:HGNC:16731] | 597 | 3.655 | -0.0765 | No |
| 8 | FAS | Fas cell surface death receptor [Source:HGNC Symbol;Acc:HGNC:11920] | 689 | 3.502 | -0.1016 | No |
| 9 | KCNN4 | potassium calcium-activated channel subfamily N member 4 [Source:HGNC Symbol;Acc:HGNC:6293] | 842 | -3.297 | -0.1594 | No |
| 10 | KCTD12 | potassium channel tetramerization domain containing 12 [Source:HGNC Symbol;Acc:HGNC:14678] | 1139 | -3.751 | -0.2886 | No |
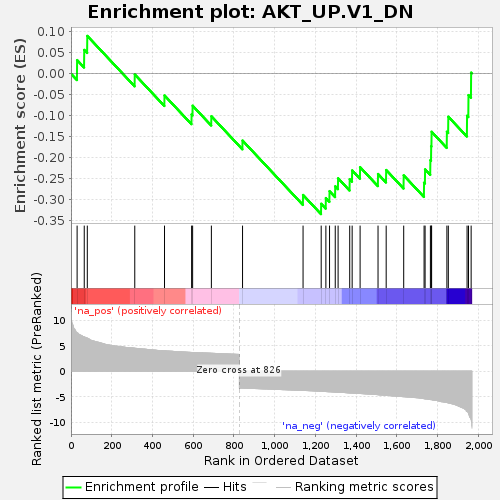
| 11 | HTRA1 | HtrA serine peptidase 1 [Source:HGNC Symbol;Acc:HGNC:9476] | 1228 | -3.913 | -0.3095 | Yes |
| 12 | ZNF385A | zinc finger protein 385A [Source:HGNC Symbol;Acc:HGNC:17521] | 1251 | -3.976 | -0.2960 | Yes |
| 13 | GRAMD1A | GRAM domain containing 1A [Source:HGNC Symbol;Acc:HGNC:29305] | 1269 | -4.014 | -0.2797 | Yes |
| 14 | NCAM1 | neural cell adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:7656] | 1297 | -4.085 | -0.2680 | Yes |
| 15 | CD248 | CD248 molecule [Source:HGNC Symbol;Acc:HGNC:18219] | 1311 | -4.114 | -0.2490 | Yes |
| 16 | SASH1 | SAM and SH3 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:19182] | 1368 | -4.249 | -0.2513 | Yes |
| 17 | PDE9A | phosphodiesterase 9A [Source:HGNC Symbol;Acc:HGNC:8795] | 1380 | -4.269 | -0.2303 | Yes |
| 18 | SMARCD3 | "SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 [Source:HGNC Symbol;Acc:HGNC:11108]" | 1419 | -4.362 | -0.2226 | Yes |
| 19 | PDIA5 | protein disulfide isomerase family A member 5 [Source:HGNC Symbol;Acc:HGNC:24811] | 1507 | -4.586 | -0.2388 | Yes |
| 20 | CDC42BPA | CDC42 binding protein kinase alpha [Source:HGNC Symbol;Acc:HGNC:1737] | 1547 | -4.728 | -0.2293 | Yes |
| 21 | IRS2 | insulin receptor substrate 2 [Source:HGNC Symbol;Acc:HGNC:6126] | 1633 | -4.979 | -0.2420 | Yes |
| 22 | ID3 | "inhibitor of DNA binding 3, HLH protein [Source:HGNC Symbol;Acc:HGNC:5362]" | 1733 | -5.369 | -0.2595 | Yes |
| 23 | ZEB1 | zinc finger E-box binding homeobox 1 [Source:HGNC Symbol;Acc:HGNC:11642] | 1738 | -5.405 | -0.2277 | Yes |
| 24 | ACKR3 | atypical chemokine receptor 3 [Source:HGNC Symbol;Acc:HGNC:23692] | 1764 | -5.541 | -0.2060 | Yes |
| 25 | IL13RA1 | interleukin 13 receptor subunit alpha 1 [Source:HGNC Symbol;Acc:HGNC:5974] | 1768 | -5.559 | -0.1727 | Yes |
| 26 | PAQR7 | progestin and adipoQ receptor family member 7 [Source:HGNC Symbol;Acc:HGNC:23146] | 1770 | -5.563 | -0.1384 | Yes |
| 27 | S1PR3 | sphingosine-1-phosphate receptor 3 [Source:HGNC Symbol;Acc:HGNC:3167] | 1845 | -6.123 | -0.1383 | Yes |
| 28 | AXIN2 | axin 2 [Source:HGNC Symbol;Acc:HGNC:904] | 1852 | -6.185 | -0.1027 | Yes |
| 29 | PDE4B | phosphodiesterase 4B [Source:HGNC Symbol;Acc:HGNC:8781] | 1944 | -7.880 | -0.1003 | Yes |
| 30 | AMPD2 | adenosine monophosphate deaminase 2 [Source:HGNC Symbol;Acc:HGNC:469] | 1951 | -8.323 | -0.0513 | Yes |
| 31 | BEX4 | brain expressed X-linked 4 [Source:HGNC Symbol;Acc:HGNC:25475] | 1964 | -9.522 | 0.0021 | Yes |
Table: GSEA details [plain text format]