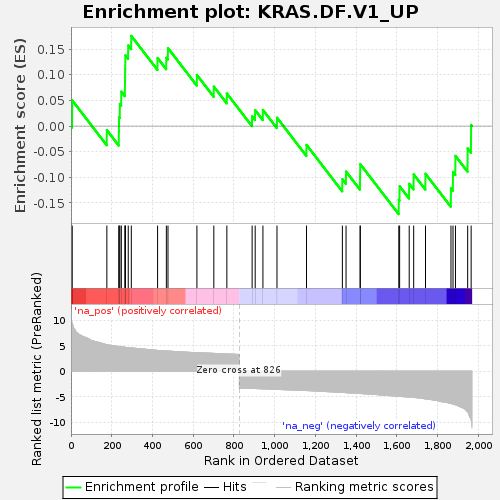
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | IRF4 | interferon regulatory factor 4 [Source:HGNC Symbol;Acc:HGNC:6119] | 6 | 9.385 | 0.0501 | Yes |
| 2 | IFITM1 | interferon induced transmembrane protein 1 [Source:HGNC Symbol;Acc:HGNC:5412] | 176 | 5.188 | -0.0079 | Yes |
| 3 | BORA | BORA aurora kinase A activator [Source:HGNC Symbol;Acc:HGNC:24724] | 235 | 4.850 | -0.0104 | Yes |
| 4 | CRYZ | crystallin zeta [Source:HGNC Symbol;Acc:HGNC:2419] | 236 | 4.850 | 0.0171 | Yes |
| 5 | ARHGEF3 | Rho guanine nucleotide exchange factor 3 [Source:HGNC Symbol;Acc:HGNC:683] | 240 | 4.829 | 0.0430 | Yes |
| 6 | ISG20 | interferon stimulated exonuclease gene 20 [Source:HGNC Symbol;Acc:HGNC:6130] | 247 | 4.810 | 0.0671 | Yes |
| 7 | SPRY2 | sprouty RTK signaling antagonist 2 [Source:HGNC Symbol;Acc:HGNC:11270] | 265 | 4.684 | 0.0849 | Yes |
| 8 | ZNF654 | zinc finger protein 654 [Source:HGNC Symbol;Acc:HGNC:25612] | 266 | 4.670 | 0.1114 | Yes |
| 9 | ATP9A | ATPase phospholipid transporting 9A (putative) [Source:HGNC Symbol;Acc:HGNC:13540] | 267 | 4.667 | 0.1379 | Yes |
| 10 | IFI27 | interferon alpha inducible protein 27 [Source:HGNC Symbol;Acc:HGNC:5397] | 281 | 4.600 | 0.1572 | Yes |
| 11 | SAP30 | Sin3A associated protein 30 [Source:HGNC Symbol;Acc:HGNC:10532] | 296 | 4.557 | 0.1758 | Yes |
| 12 | BCL2L1 | BCL2 like 1 [Source:HGNC Symbol;Acc:HGNC:992] | 425 | 4.064 | 0.1327 | No |
| 13 | HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 [Source:HGNC Symbol;Acc:HGNC:24368] | 468 | 3.952 | 0.1334 | No |
| 14 | DDB2 | damage specific DNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:2718] | 476 | 3.934 | 0.1520 | No |
| 15 | ETFB | electron transfer flavoprotein subunit beta [Source:HGNC Symbol;Acc:HGNC:3482] | 618 | 3.622 | 0.0996 | No |
| 16 | IFIT1 | interferon induced protein with tetratricopeptide repeats 1 [Source:HGNC Symbol;Acc:HGNC:5407] | 701 | 3.484 | 0.0770 | No |
| 17 | NAT1 | N-acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:7645] | 765 | 3.377 | 0.0635 | No |
| 18 | DNASE1L1 | deoxyribonuclease 1 like 1 [Source:HGNC Symbol;Acc:HGNC:2957] | 889 | -3.364 | 0.0190 | No |
| 19 | TBC1D8 | TBC1 domain family member 8 [Source:HGNC Symbol;Acc:HGNC:17791] | 904 | -3.388 | 0.0310 | No |
| 20 | RGS2 | regulator of G protein signaling 2 [Source:HGNC Symbol;Acc:HGNC:9998] | 942 | -3.447 | 0.0314 | No |
| 21 | CPVL | carboxypeptidase vitellogenic like [Source:HGNC Symbol;Acc:HGNC:14399] | 1011 | -3.552 | 0.0163 | No |
| 22 | AKAP12 | A-kinase anchoring protein 12 [Source:HGNC Symbol;Acc:HGNC:370] | 1156 | -3.781 | -0.0367 | No |
| 23 | GLRX | glutaredoxin [Source:HGNC Symbol;Acc:HGNC:4330] | 1332 | -4.145 | -0.1037 | No |
| 24 | SMARCA1 | "SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 [Source:HGNC Symbol;Acc:HGNC:11097]" | 1350 | -4.193 | -0.0887 | No |
| 25 | SMARCD3 | "SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 [Source:HGNC Symbol;Acc:HGNC:11108]" | 1419 | -4.362 | -0.0992 | No |
| 26 | TLR2 | toll like receptor 2 [Source:HGNC Symbol;Acc:HGNC:11848] | 1420 | -4.362 | -0.0744 | No |
| 27 | CCL20 | C-C motif chemokine ligand 20 [Source:HGNC Symbol;Acc:HGNC:10619] | 1609 | -4.912 | -0.1438 | No |
| 28 | AKR1C3 | aldo-keto reductase family 1 member C3 [Source:HGNC Symbol;Acc:HGNC:386] | 1613 | -4.931 | -0.1174 | No |
| 29 | IL1RL1 | interleukin 1 receptor like 1 [Source:HGNC Symbol;Acc:HGNC:5998] | 1660 | -5.050 | -0.1126 | No |
| 30 | GALNT6 | polypeptide N-acetylgalactosaminyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:4128] | 1682 | -5.138 | -0.0943 | No |
| 31 | CXCL8 | C-X-C motif chemokine ligand 8 [Source:HGNC Symbol;Acc:HGNC:6025] | 1740 | -5.411 | -0.0931 | No |
| 32 | PID1 | phosphotyrosine interaction domain containing 1 [Source:HGNC Symbol;Acc:HGNC:26084] | 1865 | -6.321 | -0.1214 | No |
| 33 | LTBP3 | latent transforming growth factor beta binding protein 3 [Source:HGNC Symbol;Acc:HGNC:6716] | 1875 | -6.411 | -0.0897 | No |
| 34 | ANPEP | "alanyl aminopeptidase, membrane [Source:HGNC Symbol;Acc:HGNC:500]" | 1887 | -6.541 | -0.0583 | No |
| 35 | VCAN | versican [Source:HGNC Symbol;Acc:HGNC:2464] | 1947 | -7.959 | -0.0437 | No |
| 36 | BEX4 | brain expressed X-linked 4 [Source:HGNC Symbol;Acc:HGNC:25475] | 1964 | -9.522 | 0.0021 | No |
Table: GSEA details [plain text format]