CHAPTER 8
RECOMBINATION OF
DNA
The previous chapter on mutation and repair of DNA dealt mainly with small changes in DNA sequence, usually single base pairs, resulting from errors in replication or damage to DNA. The DNA sequence of a chromosome can change in large segments as well, by the processes of recombination and transposition. Recombination is the production of new DNA molecule(s) from two parental DNA molecules or different segments of the same DNA molecule; this will be the topic of this chapter. Transposition is a highly specialized form of recombination in which a segment of DNA moves from one location to another, either on the same chromosome or a different chromosome; this will be discussed in the next chapter.
Types and examples of recombination
At least four types of naturally occurring recombination have been identified in living organisms (Fig. 8.1). General or homologous recombination occurs between DNA molecules of very similar sequence, such as homologous chromosomes in diploid organisms. General recombination can occur throughout the genome of diploid organisms, using one or a small number of common enzymatic pathways. This chapter will be concerned almost entirely with general recombination. Illegitimate or nonhomologous recombination occurs in regions where no large-scale sequence similarity is apparent, e.g. translocations between different chromosomes or deletions that remove several genes along a chromosome. However, when the DNA sequence at the breakpoints for these events is analyzed, short regions of sequence similarity are found in some cases. For instance, recombination between two similar genes that are several million bp apart can lead to deletion of the intervening genes in somatic cells. Site-specific recombination occurs between particular short sequences (about 12 to 24 bp) present on otherwise dissimilar parental molecules. Site-specific recombination requires a special enzymatic machinery, basically one enzyme or enzyme system for each particular site. Good examples are the systems for integration of some bacteriophage, such as l, into a bacterial chromosome and the rearrangement of immunoglobulin genes in vertebrate animals. The third type is replicative recombination, which generates a new copy of a segment of DNA. Many transposable elements use a process of replicative recombination to generate a new copy of the transposable element at a new location.
Recombinant DNA technology uses two other types of recombination. The directed cutting and rejoining of different DNA molecules in vitro using restriction endonucleases and DNA ligases is well-known, as covered in Chapter 2. Once made, these recombinant DNA molecules are then introduced into a host organism, often a bacterium. If the recombinant DNA is a plasmid, phage or other molecule capable of replicating in the host, it will stay extrachromosomal. However, one can introduce the recombinant DNA molecule into a host in which it cannot replicate, such as a plant, an animal cell in culture, or a fertilized mouse egg. In order for the host to be stably transformed, the introduced DNA has to be taken up into a host chromosome. In bacteria and yeast, this can occur by homologous recombination at a reasonably high frequency. However, this does not occur in plant or animal cells. In contrast, at a low frequency, some of these introduced DNA molecules are incorporated into random locations in the chromosomes of the host cell. Thus random recombination into chromosomes can make stably transfected cells and transgenic plants and animals. The mechanism of this recombination during transformation or transfection is not well understood, although it is commonly used in the laboratory.
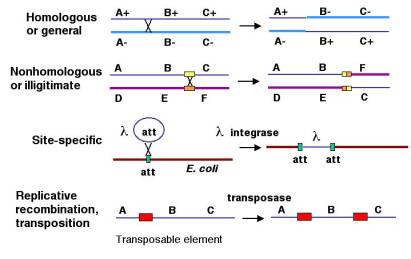
Figure 8.1. Types of natural recombination. Each line represents a chromosome or segment of a
chromosome; thus a single line represents both strands of duplex DNA. For homologous
or general recombination, each homologous
chromosome is shown as a different shade of blue and a distinctive thickness,
with different alleles for each of the three genes on each. Recombination
between genes A and B leads to a reciprocal exchange of genetic information,
changing the arrangement of alleles on the chromosomes. For nonhomologous
(or illegitimate) recombination, two
different chromosomes (denoted by the different colors and different genes)
recombine, moving, e.g. gene C so that it is now on the same chromosome as
genes D and E. Although the sequences of the two chromosomes differ for most of
their lengths, the segments at the sites of recombination may be related,
denoted by the yellow and orange rectangles. Site-specific
recombination leads to the combination of
two different DNA molecules, illustrated here for a bacteriophage l integrating into the E. coli chromosome. This reaction is catalyzed by a specific
enzyme that recognizes a short sequence present in both the phage DNA and the
target site in the bacterial chromosome, called att. Replicative recombination is seen for
some transposable elements, shown as red rectangles, again using a specific
enzyme, in this case encoded by the transposable element.
General recombination is an integral part of the complex process of meiosis in sexually reproducing organisms. It results in a crossing over between pairs of genes along a chromosome, which are revealed in appropriate matings (Chapter 1). The chiasmata that link homologous chromosomes during meiosis are the likely sites of the crossovers that result in recombination. General recombination also occurs in nonsexual organisms when two copies of a chromosome or chromosomal segment are present. We have encountered this as recombination during F-factor mediated conjugal transfer of parts of chromosomes in E. coli (Chapter 1). Recombination between two phage during a mixed infection of bacteria is another example. Also, the retrieval system for post-replicative repair (Chapter 7) involves general recombination.
The mechanism of recombination has been intensively studied in bacteria and fungi, and some of the enzymes involved have been well characterized. However, a full picture of the mechanism, or mechanisms, of recombination has yet to be achieved. We will discuss the general properties of recombination, cover two models of recombination, and discuss some of the properties of key enzymes in the pathways of recombination.
Reciprocal and nonreciprocal recombination
General recombination can appear to result in either an equal or an unequal exchange of genetic information. Equal exchange is referred to as reciprocal recombination, as illustrated in Fig. 8.1. In this example, two homologous chromosomes are distinguished by having wild type alleles on one chromosome (A+, B+ and C+) and mutant alleles on the other (A-, B- and C-). Homologous recombination between genes A and B exchanges the segment of one chromosome containing the wild type alleles of genes B and C (B+ and C+) for the segment containing the mutant alleles (B- and C-) on the homologous chromosome. This could be explained by breaking and rejoining of the two homologous chromosomes during meiosis; however, we will see later that the enzymatic mechanism is more complex than simple cutting and ligation. The DNA that is removed from the top (thin dark blue) chromosome is joined with the bottom (thick light blue) chromosome, and the DNA removed from the bottom chromosome is added to the top chromosome. This process resulting in new DNA molecules that carry genetic information derived from both parental DNA molecules is called reciprocal recombination. The number of alleles for each gene remains the same in the products of this recombination, only their arrangement has changed.
General
recombination can also result in a one-way transfer of genetic information,
resulting in an allele of a gene on one chromosome being changed to the allele
on the homologous chromosome. This is called gene conversion. As illustrated in Fig. 8.2, recombination between
two homologous chromosomes A+B+C+ and A-B-C- can result in a new arrangement,
A-B+C-, without a change in the parental A+B+C+. In this case, the allele of
gene B on the bottom chromosome has changed from B- to B+ without a reciprocal
change on the other chromosome. Thus, in contrast to reciprocal recombination,
the number of types of alleles for gene B has changed in the products of this
recombination; now there is only one (B+). This is an example of
interchromosomal gene conversion, i.e. between homologous chromosomes. Similar
copies of genes can be on the same chromosome, and these can undergo gene
conversion as well. Cases of intrachromosomal gene conversion have been
documented for the gamma-globin genes of humans. The occurrence of gene
conversion during general recombination is one indication that the enzymatic
mechanism is not a simple cutting and pasting.
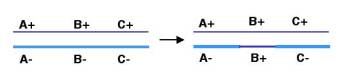
Figure 8.2. Gene conversion changing
allele B- on the bottom (thick, light blue) chromosome to B+. Note that the
arrangement of alleles on the top (thin, dark blue) chromosome has not changed.
Question 8.1. Why would you not interpret the A-B+C- chromosome as resulting from two reciprocal crossovers, one on each side of gene B?
Detecting recombination
As reviewed in Chapter 1, Mendel’s Second Law described the random assortment of alleles of pairs of genes. However, certain pairs of genes show deviations from this random assortment, leading to the conclusion that those genes are linked on a chromosome. The linkage is not always complete, meaning that nonparental genotypes are seen in a proportion of the progeny. This is explained by crossing over between the gene pairs during meiosis in the parents.
Let’s think about the general recombination shown in Fig. 8.1 in this context. The two chromosomes outlined in the figure are in a heterozygous parent, with the wild type alleles for genes A and B (A+ and B+) are on one chromosome and the mutant alleles (A- and B-) are on the homologous chromosome (We can ignore gene C for this discussion.) Homologous recombination during meiosis can generate the new chromosomes shown, now with A+ and B- on one chromosome and A- and B+ on the other. However, this crossover will not occur between genes A and B on all chromosomes undergoing meiosis in this parent. Although recombination is an essential part of meiosis (see next section), the sites of recombination on a particular chromosome varies from cell to cell. In fact, the probability that a crossover will occur between two genes is a measure of the genetic distance between them (reviewed in Chapter 1). The recombinant chromosomes resulting from a crossover are revealed in a mating between the heterozygous parent (A+B+/A-B-) and a homozygous recessive individual (A-B-/A-B-). Most of the germ cells contributed by the heterozygous parent will have one of the parental chromosomes A+B+ or A-B-, but those germ cells resulting from the crossover between genes A and B will have the recombinant chromosomes (either A+B- or A-B+). The homozygous recessive parent will contribute only A-B- chromosomes. Thus in the progeny, one sees mainly offspring whose phenotype is determined by one of the chromosomes in the heterozygous parent, either wild type A and B (genotype of A+B+/A-B-) or mutant A and B (genotype A-B-/A-B-). However, some of the progeny will show a wild type A and a mutant B phenotype, or vice versa. These carry the chromosomes resulting from the crossover (genotype of A+B-/A-B- or A-B+/A-B-). The frequency with which one sees progeny with nonparental phenotypes is related to their distance apart on the chromosome; this measure is referred to as a genetic distance or a recombination distance.
Meiotic recombination
A diploid organism has two copies of each chromosome. If it has four chromosomes, there are two pairs, A and A’ and B and B’, not four different chromosomes A, B, C and D. One copy of each chromosome came from its father (e.g. A and B) and one copy of each came from its mother (e.g. A’ and B’). Meiosis is the process of reductive division whereby a diploid organism generates haploid germ cells (in this case, with two chromosomes), and each germ cell has a single copy of each chromosome. In this example, meiosis does not generate germ cells with A and A’ or B and B’, rather it produces cells with A and B, or A and B’, or A’ and B, or A’ and B’. The homologous chromosomes, each consisting of two sister chromatids, are paired during the first phase of meiosis, e.g., A with A’ and B with B’ (Fig. 8.3; see also Figs. 1.3 and 1.4). Then the homologous chromosomes are moved to separate cells at the end of the first phase, insuring that the two homologs do not stay together during reductive division in the second phase of meiosis. Thus each germ cell receives the haploid complement of the genetic material, i.e. one copy of each chromosome. The combination of two haploid sets of chromosomes during fertilization restores the diploid state, and the cycle can resume. Failure to distribute one copy of each chromosome to each germ cell has severe consequences. Absence of one copy of a chromosome in an otherwise diploid zygote is likely fatal. Having an extra copy of a chromosome (trisomy) also causes problems. In humans, trisomy for chromosomes 15 or 18 results in perinatal death and trisomy 21 leads to developmental defects known as Down’s syndrome.
Question 8.2. If this diploid organism with chromosomes A, A’, B and B’ underwent meiosis without homologous pairing and separation of the homologs to different cells, what fraction of the resulting haploid cells would have an A-type chromosome (A or A’) and a B-type chromosome (B or B’)?
The ability of homologous chromosomes to be paired during the first phase of meiosis is fundamental to the success of this process, which maintains a correct haploid set of chromosomes in the germ cell. Recombination is an integral part of the pairing of homologous chromosomes. It occurs between non-sister chromatids during the pachytene stage of meiosis I (the first stage of meiosis) and possibly before, when the homologous chromosomes are aligned in zygotene (Fig. 8.3). The crossovers of recombination are visible in the diplotene phase. During this phase, the homologous chromosomes partially separate, but they are still held together at joints called chiasmata; these are likely the actual crossovers between chromatids of homologous chromosomes. The chiasmata are progressively broken as meiosis I is completed, corresponding to resolution of the recombination intermediates. During anaphase and telophase of meiosis I, each homologous chromosome moves to a different cell, i.e. A and A’ in different cells, B and B’ in different cells in our example. Thus recombinations occur in every meiosis, resulting in at least one exchange between pairs of homologous chromosomes per meiosis.
Recent genetic evidence
demonstrates that recombination is required for homologous pairing of
chromosomes during meiosis. Genetic screens have revealed mutants of yeast and Drosophila that block pairing of
homologous chromosomes. These are also defective in recombination. Likewise,
mutants defective in some aspects of recombination are also defective in pairing.
Indeed, the process of synapsis (or pairing) between homologous chromosomes in
zygotene, crossing over between homologs in pachytene, and resolution of the
crossovers in the latter phases of meiosis I (diakinesis, metaphase I and
anaphase I) correspond to the synapsis, formation of a recombinant joint and
resolution that mark the progression of recombination, as will be explained
below.
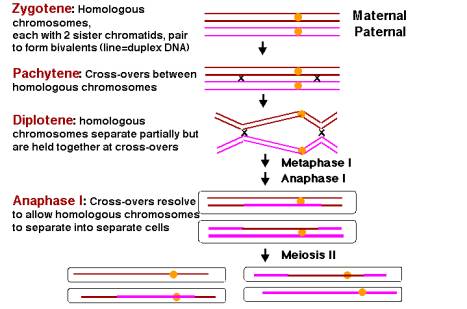
Figure 8.3.
Homologous pairing and recombination during the first stage of meiosis (meiosis
I). After DNA synthesis has been completed, two copies of each homologous
chromosome are still connected at centromeres (yellow circles). This diagram
starts with replicated chromosomes, referred to as the four-strand stage in the
literature on meiosis and recombination. In this usage, each “strand” is a chromatid and is a duplex DNA molecule. In this diagram, each
duplex DNA molecule is shown as a single line, brown for the two sister
chromatids of chromosome derived from the mother (maternal) and pink for the
sister chromatids from the paternal chromosome. Only one homologous pair is
shown, but ususally there are many more, e.g. 4 pairs of chromosomes in Drosophila and 23 pairs in humans. During the meiosis I, the
homologous chromosomes align and then separate. At the zygotene stage, the two
homologous chromosomes, each with two sister chromatids, pair along their
length in a process called synapsis. The resulting group of four chromatids is
called a tetrad or bivalent. During pachytene, recombination occurs between a
maternal and a paternal chromatid, forming crossovers between the homologous
chromosomes. The two homologous chromosomes separate along much of their length
at diplotene, but they continue to be held together at localized chiasmata,
which appear as X-shaped structures in micrographs. These physical links are
thought to be the positions of crossing over. During metaphase and anaphase of
the first meiotic division, the crossovers are gradually broken (with those at
the ends resolved last) and the two homologous chromosomes (each still with two
chromatids joined at a centromere) are moved into separate cells. During the
second meiotic division (meiosis II), the centromere of each chromosome
separates, allowing the two chromatids to move to separate cells, thus
finishing the reductive division and making four haploid germ cells.
Advantages of genetic recombination
Not only is recombination needed for homologous pairing during meiosis, but recombination has at least two additional benefits for sexual species. It makes new combinations of alleles along chromosomes, and it restricts the effects of mutations largely to the region around a gene, not the whole chromosome.
Since each chromosome undergoes at least one recombination event during meiosis, new combinations of alleles are generated. The arrangement of alleles inherited from each parent are not preserved, but rather the new germ cells carry chromosomes with new combinations of alleles of the genes (Fig. 8.4). This remixing of combinations of alleles is a rich source of diversity in a population.
![]()
![]()
![]()
![]()
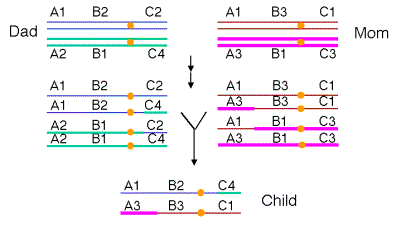
Figure 8.4. Recombination during meiosis generates new
combinations of alleles in the offspring. One homologous pair of chromosomes is
illustrated, starting at the “four-strand” stage. Each line is a duplex DNA
molecule in a chromatid. The two chromosomes in the father (inherited from the
paternal grandparents) are blue and green; the homologous chromosomes in the
mother (inherited from the maternal grandparents) are brown and pink. All
chromosomes have genes A, B and C; different numbers refer to different
alleles. In this illustration, a crossover on the short arm of the chromosome
during development of the male germ cells links allele 4 of gene C with alleles
1 of gene A and allele 2 of gene B, as well as the reciprocal arrangement. A
crossover on the long arm of the chromosome is illustrated for development of
the female germ cell, making the new combination A3, B3 and C1. A child can
have the new chromosomes A1B2C4 and A3B3C1. Note that neither of these
combinations was in the father or mother.
Over time, recombination will separate alleles at one locus from alleles at a linked locus. A chromosome through generations is not fixed, but rather it is "fluid," having many different combinations of alleles. This allows nonfunctional (less functional) alleles to be cleared from a population. If recombination did not occur, then one deleterious mutant allele would cause an entire chromosome to be eliminated from the population. However, with recombination, the mutant allele can be separated from the other genes on that chromosome. Then negative selection can remove defective alleles of a gene from a population while affecting the frequency of alleles only of genes in tight linkage to the mutant gene. Conversely, the rare beneficial alleles of genes can be tested in a population without being irreversibly linked to any potentially deleterious mutant alleles of nearby genes. This keeps the effective target size for mutation close to that of a gene, not the whole chromosome.
Evidence for heteroduplexes from recombination in fungi
The mechanism by which recombination occurs has been studied primarily in fungi, such as the budding yeast Saccharomyces cerevisiae and the filamentous fungus Ascomycetes, and in bacteria. The fungi undergo meiosis, and hence some aspects of their recombination systems may be more similar to that of plants and animals than is that of bacteria. However, the enzymatic functions discovered by genetic and biochemical studies of recombination in bacteria are also proving to have counterparts in eukaryotic organisms as well. We will refer to studies mainly in fungi for the models of recombination, and to studies mainly in bacteria for the enzymatic pathways.
Many important insights into the mechanism of recombination have come from studies in fungi. One fundamental observation is that recombination proceeds by the formation of a region of heteroduplex, i.e. the recombination products have a region with one strand from one chromosome and the complementary strand from the other chromosome. Thus recombination is not a simple cut and paste operation, unlike the joining of two different molecules by recombinant DNA technology. The two recombining molecules are joined and form a hybrid, or heteroduplex, over part of their lengths.
The anatomy and physiology of the filamentous fungus Ascomycetes allows one to observe this heteroduplex formed during recombination. A cell undergoing meiosis starts with a 4n complement of chromosomes (i.e. twice the diploid number) and undergoes two rounds of cell division to form four haploid cells. In fungi these haploid germ cells are spores, and they are found together in an ascus. They can be separated by dissection and plated individually to examine the phenotype of the four products of meiosis. This is called tetrad analysis.
The fungus Ascomycetes goes one step further. After meiosis is completed, the germ cells undergo one further round of replication and mitosis. This separates each individual polynucleotide chain (or “strand” in the sense used in nucleic acid biochemistry) of each DNA duplex in the meiotic products into a separate spore. The eight spores in the ascus reflect the genetic composition of each of the eight polynucleotide chains in the four homologous chromosomes. (The two sister chromatids in each homologous chromosome become two chromosomes after meiosis, and each chromosome is a duplex of two polynucleotide chains.)
The order of the eight spores in the ascus of Ascomycetes reflects the descent of the spores from the homologous chromosomes. As shown in Fig. 8.5, a heterozygote with a “blue” allele on one homologous chromosome and a “red” allele on the other will normally produce four “blue” spores and four “red” spores. The four spores with the same phenotype were derived from one homologous chromosome and are adjacent to each other in the ascus. This is called a 4:4 parental ratio, i.e. with respect to the phenotypes of the parent of the heterozygote.
The evidence for heteroduplex formation comes from deviations from the normal 4:4 ratio. Sometimes a 3:5 parental ratio is seen for a particular genetic marker. This shows that one polynucleotide chain of one allele has been lost (giving 4-1=3 spores with the corresponding phenotype in the ascus) and replaced by the polynucleotide chain of the other allele (giving 4+1=5 spores with the corresponding phenotype). As illustrated in Fig. 8.5, this is 3 blue spores and 5 red spores. The segment of the chromosome containing this gene was a heteroduplex with one chain from each of two alleles. The round of replication and mitosis that follows meiosis in this fungus allows the two chains to be separated into two alleles that generated a different phenotype in a plating assay. Thus this 3:5 ratio results from post-meiotic segregation of the two chains of the different alleles. In this fungus, a region of heteroduplex can be directly observed by a plating assay.
The region of heteroduplex is associated with a recombination between the chromosomes. Other genes flank the region of heteroduplex shown in Fig. 8.5. In many cases, the arrangement of alleles of these flanking genes has changed from that on the parental chromosomes, reflecting a recombination. For instance, let the region of heteroduplex be in a gene B, flanked by gene A in the left and gene C on the right. Each gene has a blue allele and a red allele, making the parental chromosomes AbBbCb and ArBrCr. If one monitored the phenotypes of determined by genes A and C (in addition to B) in the third and fourth spores (derived from the chromosome with the heteroduplex), they would see the phenotypes for the nonparental chromosomes AbBbCr and AbBrCr. This change in the flanking markers (genes A and C) reflects a recombination. Thus the heteroduplex can be found between markers that have undergone recombination.
Other
markers can show a 2:6 parental ratio. This means that one of the alleles
(formerly blue in fig. 8.5) has been changed to the other allele (now red), in
a process called gene conversion. This
can occur between flanking markers that have been switched because of
recombination. Thus like the heteroduplex, the region of gene conversion is
associated with recombination. Models for recombination need to incorporate
both phenomenon into their proposed mechanism.
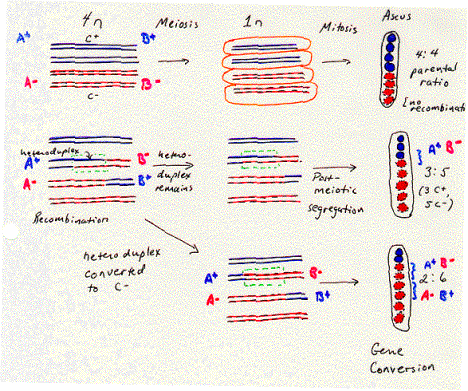
Figure 8.5. Spores formed during meiosis in Ascomycetes reflect the genetic composition of the parental DNA chains. The four homologous chromosomes in the 4n state are shown as duplex DNA molecules, with one line for each DNA chain. Two sister chromatids are blue and two sister chromatids are red, reflecting their ability to be distinguished in a plating assay for particular genes along the chromosome. Meiosis places each of the four homologous chromosomes into a different cell, and in this species, it is followed by replication and mitosis so that each of the eight spores (circles in the elongated ellipse representing the ascus) has the genetic composition of each of the eight DNA chains in the four chromosomes that result from meiosis (two complementary chains per chromosome). A region of heteroduplex can be seen as a 3:5 parental ratio after post-meiotic segregation. A region of gene conversion can be seen as a 2:6 parental ratio.
Question 8.3. Imagine that you are studying a fungus that generates an ascus with 8 spores like Ascomycetes, in which the products of meiosis complete an additional round of replication and mitosis. You generate a heterozyous strain by mating a parent that was homozyous for the markers leu+, SmR, ade8+ and another that was leu-, SmS, ade8-. Previous studies had shown that all three markers are linked in the order given. Each of these pairs of alleles can be distinguished in a plating assay. The allele leu+ confers leucine auxotrophy whereas leu- confers leucine prototrophy. The allele SmR confers resistance to spectinomycin whereas SmS is sensitive to this antibiotic. Colonies of fungi with the ade8+ allele give a red color in under appropriate conditions in a plate, but those with the ade8- are white. Analysis of the individual spores from an ascus gave the following phenotypes results. The spores are numbered in the order they were in the ascus. What are the corresponding genotypes of the chromosome in each spore? How do you interpret these results with respect to recombination?
|
Spore |
leucine |
Spectinomycin |
Color in ade test |
|
1 |
prototroph |
resistant |
red |
|
2 |
prototroph |
resistant |
red |
|
3 |
prototroph |
resistant |
white |
|
4 |
prototroph |
sensitive |
white |
|
5 |
auxotroph |
sensitive |
red |
|
6 |
auxotroph |
sensitive |
red |
|
7 |
auxotroph |
sensitive |
white |
|
8 |
auxotroph |
sensitive |
white |
Holliday model for general recombination: Single strand
invasion
In 1964, Robin Holliday proposed a model that accounted for heteroduplex formation and gene conversion during recombination. Although it has been supplanted by the double-strand break model (at least for recombination in yeast and higher organisms), it is a useful place to start. It illustrates the critical steps of pairing of homologous duplexes, formation of a heteroduplex, formation of the recombination joint, branch migration and resolution.
The steps in the Holliday Model are illustrated in Fig. 8.6.
(1) Two homologous chromosomes, each composed of duplex DNA, are paired with similar sequences adjacent to each other.
(2) An endonuclease nicks at corresponding regions of homologous strands of the paired duplexes. This is shown for the strands with the arrow to the right in the figure.
(3) The nicked ends dissociate from their complementary strands and each single strand invades the other duplex. This occurs in a reciprocal manner to produce a heteroduplex region derived from one strand from each parental duplex.
(4) DNA ligase seals the nicks. The result is a stable joint molecule, in which one strand of each parental duplex crosses over into the other duplex. This X-shaped joint is called a Holliday intermediate or Chi structure.
(5) Branch migration then expands the region of heteroduplex. The stable joint can move along the paired duplexes, feeding in more of each invading strand and extending the region of heteroduplex.
(6) The recombination intermediate is then resolved by nicking a strand in each duplex and ligation.
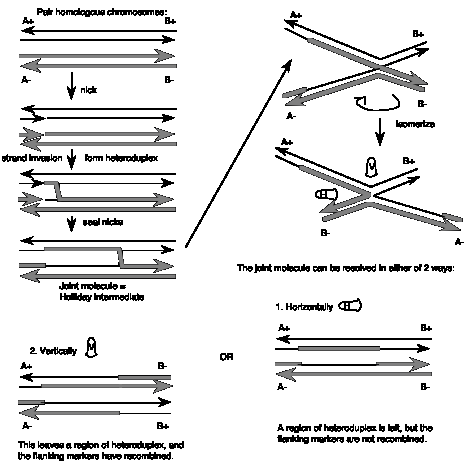
Figure 8.6. Holliday model for general recombination: Single strand invasion. Each of the polynucleotide chains (or strands of the duplex) is shown with a particular orientation, indicated by the arrows. The chromosomes with thick chains and thin chains are homologous. The chains closest to each other in this diagram of the homologous chromosomes are shown in the same orientation. (In contrast to many of the figures in this book, the top strand of each duplex is not necessarily oriented 5’ to 3’ left to right.) The Holliday model does not specify a particular end (5’ or 3’) for the invading single strand, but for ease in following the events, the ends are given an orientation in the figure.
Resolution can occur in either of two ways, only one of which results in an exchange of flanking markers after recombination. The two modes of resolution can be visualized by rotating the duplexes so that no strands cross over each other in the illustration (Fig. 8.6). In the “horizontal” mode of resolution, the nicks are made in the same DNA strands that were originally nicked in the parental duplexes. After ligation of the two ends, this produces two duplex molecules with a patch of heteroduplex, but no recombination of flanking regions. In contrast, for the “vertical” mode of resolution, the nicks are made in the other strands, i.e. those not nicked in the original parental duplexes. Ligation of these two ends also leaves a patch of heteroduplex, but additionally causes recombination of flanking regions. Note that “horizontal” and “vertical” are just convenient designations for the two modes based on the two-dimensional drawings that we can make. The important distinction in terms of genetic outcome is whether the resolution steps target the strands initially cleaved or the other strand.
The steps in this model of general recombination can be viewed in a dynamic form by visiting a web site maintained by geneticists at the University of Wisconsin (URL is http://www.wisc.edu/genetics/Holliday/index.html). This shows the steps in the Holliday model in a movie, illustrating the actions much more vividly than static diagrams.
The
recombinant joint proposed by Holliday has been visualized in electron
micrographs of recombining DNA duplexes (Fig. 8.7A). It has the proposed X
shape. {This would be a good place to add an EM photo.} Although this joint is drawn with some distance
between the duplexes in illustrations, in fact the two duplexes are juxtaposed,
and only a very few base pairs are
broken in the Holliday intermediate (Fig. 8.7B). The structure is symmetrical ,
and it is likely that the choice between “horizontal” and “vertical” resolution
is a random event by the resolving nuclease. It chooses two strands, but it
cannot tell which were initially cleaved and which were not.
A. Add a figure here, need to find an EM picture.
B. 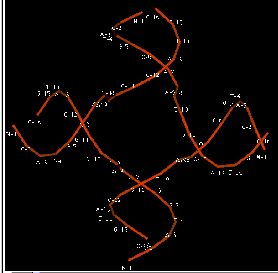
Figure 8.7. Views of a Holliday junction. A. Electron micrograph of two DNA duplexes in a recombination intermediate. B. Holliday junction from X-ray crystallography of a RuvA-Holliday junction complex (from Hargreaves et al. (1998) Nature Structural Biology 5: 441-4460. For this view, the RuvA protein tetramer was removed and only the phosphodiester backbones of the two duplexes (four strands) are shown. Note the kinks in the DNA in the center of the structure. These correspond to about three nucleotides in each strand that are not paired as in B form DNA. The atomic coordinates were downloaded from the Molecular Structure database at NCBI and rendered in Cn3D v.3.0. The positions of each nucleotide in the four strands are labeled, with a letter for the nucleotide and the number along the chain. Files for viewing the virtual 3-D image on your own computer are accessible at the course web site.
Studies of recombination between chromosomes with limited homology have shown that the minimum length of the region required to establish the connection between the recombining duplexes is about 75 bp. If the homology region is shorter than this, the rate of recombination is substantially reduced.
The patch of heteroduplex can be replicated (Fig. 8.8) or repaired to generate a gene conversion event. As shown in Fig. 8.8, replication through the products of horizontal resolution (from Fig. 8.6) will generate a duplex from each strand of the heteroduplex. If we consider the parental chromosomes to be A+C+B+ and A-C-B-, and the heteroduplex to be in gene C, the products of replication can have a the parental C+ converted to a C- but still flanked by A+ and B+ or C- converted to C+ but still flanked by A- and B-. In either case, gene C has changed to a new allele without affecting the flanking markers.
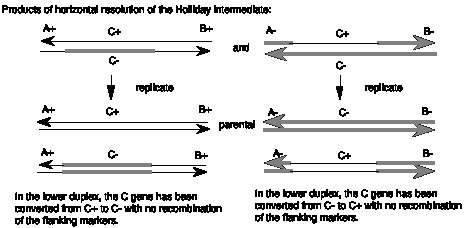
Figure 8.8. Gene conversion can occur by replication through the heteroduplex region.
Although the original Holliday model accounted for many important aspects of recombination (all that were known at the time), some additional information requires changes to the model. For instance, the Holliday model treats both duplexes equally; both are the invader and the target of the strand invasion. Also, no new DNA synthesis is required in the Holliday model. However, subsequent work showed that one of the duplex molecules is the used preferentially as the donor of genetic information. Hence additional models, such as one from Meselson and Radding, incorporated new DNA synthesis at the site of the nick to make and degradation of a strand of the other duplex to generate asymmetry into the two duplexes, with one the donor the other the recipient of DNA. These ideas and others have been incorporated into a new model of recombination involving double strand breaks in the DNAs.
Double-strand-break model for recombination
Several lines of evidence, primarily from studies of recombination in yeast, required changes to the reciprocal exchange of DNA chains initiated at single-strand nicks. As just mentioned, one DNA duplex tended to be the donor of information and the other the recipient, in contrast to the equal exchange predicted by the original Holliday model. Also, in yeast, recombination could be initiated by double-strand breaks. For instance, both DNA strands are cleaved (by the HO endonuclease) to initiate recombination between the MAT and HML(R) loci in mating type switching in yeast. Using plasmids transformed into yeast, it was shown that a double-strand gap in the “aggressor” duplex could be used to initiate recombination, and the gap was repaired during the recombination (this experiment is explored in problem 8.___). In this case, the gap in one duplex was filled by DNA donated from the other substrate. All this evidence was incorporated into a major new model for recombination from Jack Szostak and colleagues in 1983. It is called the double-strand-break model. New features in this model (contrasting with the Holliday model) are initiation at double-strand breaks, nuclease digestion of the aggressor duplex, new synthesis and gap repair. However, the fundamental Holliday junction, branch migration and resolution are retained, albeit with somewhat greater complexity because of the additional numbers of Holliday junctions. Although many aspects of the recombination mechanism differ
The steps in the double-strand-break model up to the formation of the joint molecules are diagrammed in Fig. 8.9.
(1) An endonuclease cleaves both strands of one of the homologous DNA duplexes, shown as thin blue lines in Fig. 8.9. This is the aggressor duplex, since it initiates the recombination. It is also the recipient of genetic information, as will be apparent as we go through the model.
(2) The cut is enlarged by an exonuclease to generate a gap with 3' single-stranded termini on the strands.
(3) One of the free 3' ends invades a homologous region on the other duplex (shown as thick red lines), called the donor duplex. The formation of heteroduplex also generates a D-loop (a displacement loop), in which one strand of the donor duplex is displaced.
(4) The D-loop is extended as a result of repair synthesis primed by the invading 3' end. The D-loop eventually gets large enough to cover the entire gap on the aggressor duplex, i.e. the one initially cleaved by the endonuclease. The newly synthesized DNA uses the DNA from the invaded DNA duplex (thick red line) as the template, so the new DNA has the sequence specified by the invaded DNA.
(5) When the displaced strand from the donor (red) extends as far as the other side of the gap on the recipient (thin blue), it will anneal with the other 3' single stranded end at that end of the gap. The displaced strand has now filled the gap on the aggressor duplex, donating its sequence to the duplex that was initially cleaved. Repair synthesis catalyzed by DNA polymerase converts the donor D-loop to duplex DNA. During steps 4 and 5, the duplex that was initially invaded serves as the donor duplex; i.e. it provides genetic information during this phase of repair synthesis. Conversely, the aggressor duplex is the recipient of genetic information. Note that the single strand invasion models predict the opposite, where the initial invading strand is the donor of the genetic information.
(6) DNA ligase will seal the nicks, one on the left side of the diagram in Fig. 8.9 and the other on the right side. Although the latter is between a strand on the bottom duplex and a strand on the top duplex, it is equivalent to the ligation in the first nick (the apparent physical separation is an artifact of the drawing). In both cases, sealing the nick forms a Holliday junction.
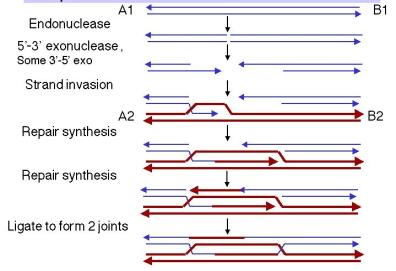
Figure 8.9. Steps in the double-strand-break model for recombination, from initiation to formation of the recombinant joints.
At this point, the recombination intermediate has two recombinant joints (Holliday junctions). The original gap in the aggressor duplex has been filled with DNA donated by the invaded duplex. The filled gap is now flanked by heteroduplex. The heteroduplexes are arranged asymmetrically, with one to the left of the filled gap on the aggressor duplex and one to the right of the filled gap on the donor duplex. Branch migration can extend the regions of heteroduplex from each Holliday junction.
The recombination intermediate can now be resolved. The presence of two recombination joints adds some complexity, but the process is essentially the same as discussed for the Holliday model. Each joint can be resolved horizontally or vertically. The key factor is whether the joints are resolved in the same mode or sense (both horizontally or both vertically) or in different modes.
If both joints are resolved the same sense (Fig. 8.10), the original duplexes will be released, each with a region of altered genetic information that is a "footprint" of the exchange event. That region of altered information is the original gap, plus or minus the regions covered by branch migration. For instance, if both joints are resolved by cutting the originally cleaved strands ("horizontally" in our diagram of the Holliday model), then you have no crossover at either joint. If both joints are resolved by cleaving the strands not cut originally ("vertically" in our diagram of the Holliday model), then you have a crossover at both joints. This closely spaced double crossover will produce no recombination of flanking markers.
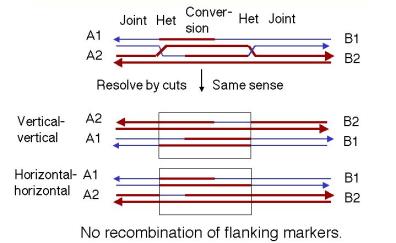
Figure 8.10. Resolution of intermediates in the
double-strand-break model by cutting the recombinant joints in the same mode or
sense. The box outlines the region between the two resolved junctions.
In contrast, if each joint is resolved in opposite directions (Fig. 8.11), then there will be recombination between flanking markers. That is, one joint will not give a crossover and the other one will.
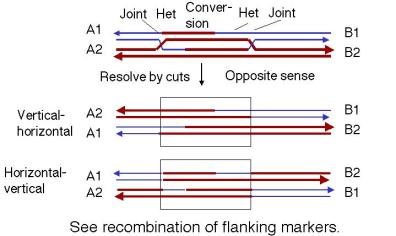
Figure 8.11. Resolution of intermediates in the double-strand-break model by cutting the recombinant joints in the opposite mode or sense.
Several features distinguish the double-strand-break model from the single-strand nick model initially proposed by Holliday. In the double-strand-break model, the region corresponding to the original gap now has the sequence of the donor duplex in both molecules. This is flanked by heteroduplexes at each end, one on each duplex. Hence the arrangement of heteroduplex is asymmetric; i.e. there is a different heteroduplex in each duplex molecule. Part of one duplex molecule has been converted to the sequence of the other (the recipient, initiating duplex has been converted to the sequence of the donor). In the single strand invasion model, each DNA duplex has heteroduplex material covering the region from the initial site of exchange to the migrating branch, i.e. the heteroduplexes are symmetric. In variations of the model (Meselson-Radding) in which some DNA is degraded and re‑synthesized, the initiating chromosome is the donor of the genetic information.
These models also have many important features in common. Steps that are common to all the models include the generation of a single strand of DNA at an end, a search for homology, strand invasion or strand exchange to form a joint molecule, branch migration, and resolution. Enzymes catalyzing each of these steps have been isolated and characterized. This is the topic of the rest of this chapter.
Enzymes required for recombination in E. coli
The initial steps in finding enzymes that carry out recombination were genetic screens for mutants of E. coli that are defective in recombination. Assays were developed to test for recombination, and mutants that showed a decrease in recombination frequency were isolated. These were assigned to complementation groups called recA, recB, recC, recD, and so forth. Roughly 20 different genes (different rec complementation groups) have been identified in E. coli. Each gene encodes an enzyme or enzyme subunit required for recombination.
Many of these genes have been cloned and their encoded products characterized in terms of a variety of enzymatic functions. However, we still do not have a clear picture of how all these enzymes work together to carry out recombination, nor has recombination has been reconstituted in vitro from purified components. Further complicating matters is the presence of multiple pathways for recombination. Much work remains to be done to completely understand recombination at a biochemical level. Despite this, the array of recombination enzymes gives us at least a partial view of the mechanisms of recombination. Also, the enzymes characterized in E. coli have homologs and counterparts in other species. Some aspects of the recombination machinery appear to be conserved across a wide phylogenetic range.
The major enzymatic steps are outlined in Fig. 8.12. Three different pathways have been characterized that differ in the steps used to generate the invading single strand of DNA. All three pathways use RecA for homologous pairing and strand exchange, RuvA and RuvB for branch migration, and RuvC and DNA ligase for resolution. These steps and enzymes will be considered individually in the following sections.
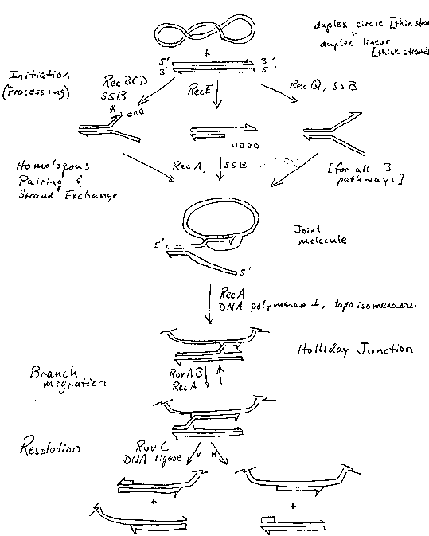
Figure 8.12. Enzymatic Steps in Recombination. Three pathways for recombination are shown, starting with a covalently closed, supercoiled circle (with each strand of the duplex shown as a thin line) and a linear duplex (with each strand shown as a thick white line) as the substrates. The three pathways differ in the enzymes used for initiation, but subsequent steps use enzymes common to all three. Adapted from Kowalczykowski, et al. (1994) Microbiological Reviews, 58:401-465.
Generation of single strands
One of the major pathways for generating 3’ single-stranded termini uses the RecBCD enzyme, also known as exonuclease V (Fig. 8.13). The three subunits of this enzyme are encoded by the genes recB, recC, and recD. Each model for recombination requires a single-strand with with a free end for strand invasion, and this enzyme does so, but with several unexpected features.
RecBCD has multiple functions, and it can switch activities. It is a helicase (in the presence of SSB), an ATPase and a nuclease. The nuclease can be a 3’ to 5’ exonuclease, and endonuclease or a 5’ to 3’ exonuclease, at different steps of the process.
The helicase activity of the RecBCD enzyme initiates unwinding only on DNA containing a free duplex end. It binds to the duplex end, using the energy of ATP hydrolysis to travel along the duplex, unwinding the DNA. The enzyme complex tracks along the top strand faster than it does on the bottom strand, so single‑stranded loops emerge, getting progressively larger as it moves down the duplex. These loops can be visualized in electron micrographs. RecBCD is also a 3' to 5' exonuclease during this phase, removing the end of one of the unwound strands (Fig. 8.13).
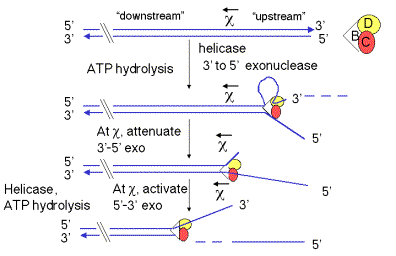
Figure 8.13. Generation of a 3'-single-stranded terminus by RecBCD enzyme
The activities of the RecBCD enzyme change at particular sequences in the DNA called chi sites (for the Greek letter c). The sequence of a chi site is 5' GCTGGTGG; this occurs about once every 4 kb on the E. coli genome. Genetic experiments show that RecBCD promotes recombination most frequently at chi sites. These sites were first discovered as mutations in bacteriophage l that led to increased recombination at those sites. These mutations altered the l sequence at the site of the mutation to become a chi site (GCTGGTGG).
When the RecBCD enzyme encounters a
chi site, it will leave an extruded single strand close to this site (4 to 6 nucleotides 3' to it). A
chi site serves as a signal to RecBCD to shift the polarity of its
exonuclease function. Before reaching the
chi site, RecBCD acts primarily as a 3’ to 5’ exonuclease, e.g. working on the
top strand in Fig. 8.13. At the chi
site, the 3’ to 5’ exonuclease function is suppressed, and after the chi
site, RecBCD converts to a 5’ to 3’ exonuclease, now working on the other
strand (e.g. the bottom strand in Fig. 8.13). Presumably, the strand that will
be the substrate for the 5’ to 3’ exonuclease is nicked in concert with this
conversion in polarity of the exonuclease. This process leaves the chi site at
the 3’ end of a single stranded DNA. This is the substrate to
which RecA can bind to initiate strand exchange (see below).
Some tests of the models for recombination have examined whether chi sites serve preferentially as either donors or recipients of the DNA during recombination. However, both results have been obtained, which makes it difficult to tie this activity precisely into either model for recombination. The genetic evidence is clear, however, that it is needed for one major pathway of recombination.
Question 8.4. What are the predictions of the Holliday model and the double-strand-break model for whether chi sites would be used as donors or recipients of genetic information during recombination?
An alternative pathway for generating single-strand ends for recombination uses the enzyme RecE, also known as exonuclease VIII. This pathway is revealed in recBCD- mutants. RecE is a 5’ to 3’ exonuclease that digests double‑stranded linear DNA, thereby generating single‑stranded 3' tails. RecE is encoded on a cryptic plasmid in E. coli. It is similar to the red exonuclease encoded by bacteriophage l.
A third pathway used the RecQ helicase, which is also a DNA-dependent ATPase. This pathway is revealed in recBCD- recE- mutants. The result of its helicase activity, in the presence of SSB, is the formation of a DNA molecule with single-stranded 3' tails, which can be used for strand invasion.
Synapsis and
invasion of single strands
The pairing of the two recombining DNA molecules (synapsis) and invasion of a single strand from the initiating duplex into the other duplex are both catalyzed by the multi-functional protein RecA. This invasion of the duplex DNA by a single stranded DNA results in the replacement of one of the strands of the original duplex with the invading strand, and the replaced strand is displaced from the duplex. Hence this reaction can also be called strand assimilation or strand exchange. RecA has many activities, including stimulating the protease function of LexA and UmuD (see Chapter 7), binding to and coating single-stranded DNA, stimulating homologous pairing between single-stranded and duplex DNA, assimilating single-stranded DNA into a duplex, and catalyzing the hydrolysis of ATP in the presence of DNA (i.e. it is a DNA-dependent ATPase). It is required in all 3 pathways for recombination. For instance, the DNA molecule with a single-stranded 3’ end generated by the RecBCD enzyme can be assimilated into a homologous region of another duplex, catalyzed by RecA and requiring the hydrolysis of ATP (Fig. 8.14).
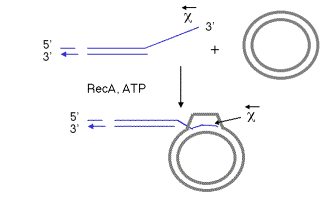
Figure 8.14. The single strand of DNA with a free 3’ end, generated by the RecBCD enzyme, can invade a homologous duplex DNA molecule in a reaction promoted by RecA. The chi site is close to the 3’ end of the single strand. The invading DNA molecule is shown with a thin, blue line for each strand. The target molecule is a duplex circle, shown as a thick gray line for each strand. ATP is required for this reaction, and it is hydrolyzed to ADP and phosphate.
The process of single-strand assimilation occurs in three steps, as illustrated in Fig. 8.15. First, RecA polymerizes onto single-stranded DNA in the presence of ATP to form the presynaptic filament. The single strand of DNA lies within a deep groove of the RecA protein, and many RecA-ATP molecules coat the single-stranded DNA. One molecule of the RecA protein covers 3 to 5 nucleotides of single-stranded DNA. The nucleotides are extended axially so they are about 5 Angstroms apart in the single-stranded DNA, about 1.5 times longer than in the absence of RecA-ATP.
Next, the presynaptic filament aligns with homologous regions in the duplex DNA. A substantial length of the three strands are held together by a polumer of RecA-ATP molecules. The aligned duplex and single strand forms a paranemic joint, meaning that the single strand is not intertwined with the double strand at this point. The duplex DNA, like the single-stranded DNA, is extended to about 1.5 times longer than in normal B form DNA (18.6 bp per turn). This extension is thought to be important in homologous pairing.
Finally, the strands are exchanged from to form a plectonemic joint. In this stage, the invading single strand is now intertwined with the complementary strand in the duplex, and one strand of the invaded duplex is now displaced. In E. coli, exchange occurs in a 5' to 3' direction relative to the single strand and requires ATP hydrolysis. In contrast, the yeast homolog, Rad51, causes the single-strand to invade with the opposite polarity, i.e. 3' to 5'. Thus the direction of this polarity is not a universally conserved feature of recombination mechanisms.
The product of strand assimilation is a heteroduplex in which one strand of the duplex was the original single-stranded DNA. The other strand of the original duplex is displaced.
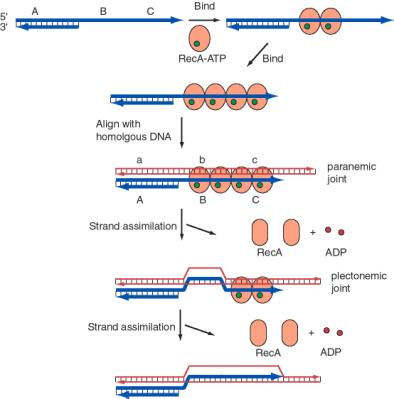
Figure 8.15. Role of RecA in assimilation of single-stranded DNA. A DNA molecule with a single-stranded 3’ end is shown with a thick blue line for each strand. A, B, and C denote particular DNA sequences. A homologous duplex is shown with thin red lines for each strand, with a, b, and c homologous to A, B and C, respectively. RecA is an orange-brown oval. It has a different conformation (shape) when ATP (green circle) is bound. The ATPase activity of RecA generates ADP (red circle) and an altered conformation of RecA, which dissociates as the single strand is assimilated. The single strand enters the duplex with a 5’ to 3’ polarity (relative to the orientation of the invading single strand).
Many details of the activity of RecA have been revealed by in vitro assays for single strand assimilation, or strand exchange. The DNA substrates for strand exchange catalyzed by RecA must meet three requirements. There must be a region of single stranded DNA on which RecA can bind and polymerize, the two molecules undergoing strand exchange must have a region of homology, and there must be a afree end within the region of homology. The latter requirement can be overcome by providing a topoisomerase.
One such assay is the conversion of a single-stranded circular DNA to a duplex circle (Fig. 8.16). The substrates for this reaction are a circular single-stranded DNA and a homologous linear duplex. These are mixed together in the presence of RecA and ATP. Many RecA-ATP molecules coat the single-stranded circle to form the nucleoprotein presynaptic filament, as discussed above. During synapsis, annealing is initiated with the 3' end of the strand complementary to the single-stranded circle. Thus the single strand invades with 5' to 3' polarity (with reference to its own polarity). Strand displacement, driven by ATP hydrolysis to dissociate the RecA, results in the formation of a nicked circle (one strand of which was the original single-stranded circle) and a linear single strand of DNA.
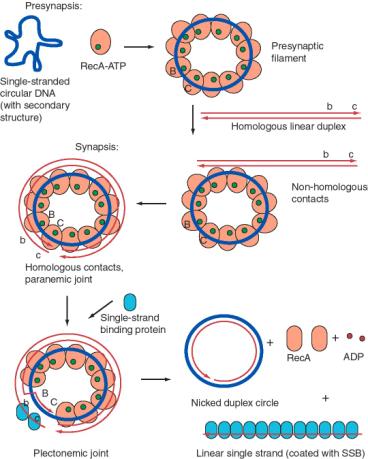
Figure 8.16. An in vitro assay for single-strand assimilation catalyzed by RecA plus ATP. Strand exchange between an invading single-stranded circle (thick blue line) and a linear duplex DNA (thin red lines), mediated by RecA plus ATP, results in a nicked duplex circle and a single-stranded linear DNA coated with single-stranded binding protein, or SSB. Regions B and C are homologous to regions b and c, respectively; they are shown as markers but the entire DNA in both molecules is homologous. SSB helps to stimulate this reaction by helping RecA overcome secondary structure in the single-stranded DNA.
Question 8.5. Try to relate this in vitro assay to the steps in the double-strand-break model for recombination. What step(s) in the model does this mimic? What else is needed for to get to the recombinant joints (Holliday junctions)?
The structure of E. coli RecA bound by ADP, both monomer and polymer, have been solved by X-ray crystallography. As shown in Fig. 8.17, the central domain has the binding site for ATP and ADP, and is presumably the site of binding of the single-stranded and double-stranded DNA. The domains extending away from the central region are involved in polymerization of RecA proteins and in interactions between the presynaptic fibers.
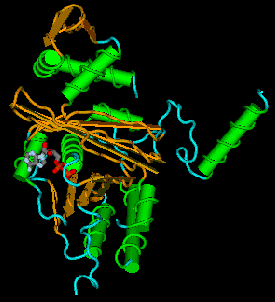
Figure 8.17. A static view of the three-dimensional structure of RecA, as determined by R. M. Story and T. A. Steitz (1992) “Structure of the recA protein-ADP complex” Nature 355: 374-376. Alpha helices are shown as green cylinders with the peptide backbone wrapped around them. Beta-sheets are yellow-brown arrows, and other regions of the peptide backbone are blue. The ADP is shown as a wire diagram, with C atoms gray, N atoms white, O atoms red and P atoms orange. Atomic coordinates were obtained from the MMDB server at NCBI and rendered in CN3D. A screen shot of one view is shown. Files for virtual 3-D viewing are available at the course web site.
A web-based tutorial showing a three-dimensional structure of RecA and illustrating aspects of its role in strand assimilation has been written by Heather M. Heerssen, Aaron Downs, and David Marcey (copyright by David Marcey). It can be accessed at the Online Museum of Macromolecules at California Lutheran University (URL is http://www.clunet.edu/BioDev/omm/reca/recamast.htm).
Proteins homologous to the E. coli RecA are found in yeast (Rad51 and Dmc1) and in mice (Rad51). Given the universality of recombination, it is likely that homologs will be found in virtually all species. Mutations in the E. coli recA gene reduce conjugational recombination by as much as 10,000 fold, so it is clear that RecA plays a central role in recombination. However, null mutations in recA are not lethal, nor are null mutations in the yeast homologs RAD51 and DMC1. In contrast, mice homozygous for a knockout mutation in the Rad51 gene die very early in development, at the 4-cell stage. This indicates that in mice, this RecA homolog is playing a novel role in replication or repair, presumably in addition to its role in recombination.
Branch migration
The movement of a Holliday junction to generate additional heteroduplex requires two proteins. One is the RuvA tetramer, which recognizes the structure of the Holliday junction. A rendering of the structure derived from X-ray crystallographic analysis of the RuvA-Holliday junction crystals is shown in Fig. 8.18.
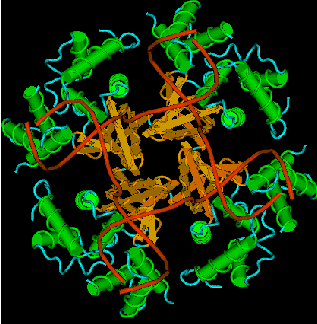
Figure 8.18. Three-dimensional structure of the RuvA tetramer complexed with a Holliday junction [from Hargreaves et al. (1998) Nature Structural Biology 5: 441-4460]. For the RuvA protein, alpha helices are green cylinders, beta sheets are brown arrows and loops are blue. The four strands of the two duplexes in the Holliday junction are red lines. The atomic coordinates were downloaded from the Molecular Structure database at NCBI, rendered in Cn3D v.3.0, and a pict file obtained as a screen shot. The kin file for viewing the virtual 3-D image on your own computer is accessible at the course web site.
RuvB is an ATPase. It forms hexameric rings that provide the motor for branch migration. As illustrated in Fig. 8.19, RuvA tetramers recognize the Holliday junction, and RuvB uses the energy of ATP hydrolysis to unwind the parental duplexes and form heteroduplexes between them.
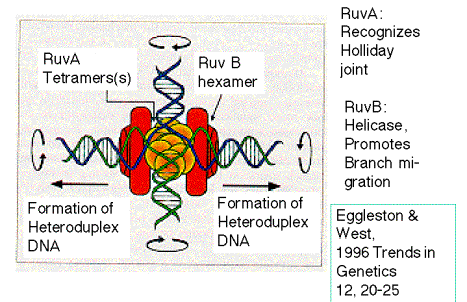
Figure 8.19. Branch migration catalyzed by RuvA and RuvB. From
Eggleston, A. K. and West, S. C. (1996) Trends in Genetics 12: 20-25.
Resolution
Ruv C is the endonuclease that cleaves the Holliday junctions (Fig. 8.20). It forms dimers that bind to the Holliday junction; recent data indicate an interaction among RuvA, RuvB and RuvC as a complex at the Holliday junction. The structure of the RuvA-Holliday junction complex (Fig. 8.18) suggests that the open structure of the junction stabilized by the binding of RuvA may expose a surface that is recognized by Ruv C for cleavage. RuvC cleaves symmetrically, in two strands with the same nearly identical sequences, thereby producing ligatable products.
The preferred site of cleavage by RuvC is 5’ WTT’S, where W = A or T and S = G or C, and ‘ is the site of cleavage. RuvC can cut strands for either horizontal or vertical resolution. Strand choice is influenced by the sequence preference and also by the presence of RecA protein, which favors vertical cleavage (i.e. to cause recombination of flanking markers).
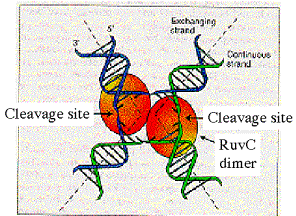
Figure 8.20.
Resolution requires cleavage by RuvC dimers.
Adapted from Eggleston, A. K. and West, S. C. (1996) Trends in Genetics 12:
20-25.
Suggested readings
Holliday, R. (1964) A mechanism for gene conversion in fungi. Genetics Research 5: 282-304.
Orr-Weaver, T. L., Szostak, J. W. and Rothstein, R. J. (1981) Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354-6358.
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. and Stahl, F. W. (1983) The double-strand-break repair model for recombination. Cell 33: 25-35.
Stahl, F. W. (1994) The Holliday junction on its thirtieth anniversary. Genetics 138: 241-246.
Kowalczykowski, S.C., Dixon, D. A., Eggleston, A. K., Lauder, S. D. and Rehrauer, W. M. (1994) Microbiological Reviews 58:401-465.
Eggleston, A. K. and West, S. C. (1996) Exchanging partners: recombination in E. coli. Treand in Genetics 12: 20-25.
Edelmann, W. and Kucherlapati, R. (1996) Role of recombination enzymes in mammalian cell survival. Proc. Natl. Acad. Sci. USA 93: 6225-6227.
Chapter 8
Recombination of
DNA
Questions
Question 8.6. According to the Holliday model for genetic recombination, what factor determines the length of the heteroduplex in the recombination intermediate?
Question 8.7. Holliday junctions can be resolved in two different ways. What are the consequences of the strand choice used I
n resolution?
Question 8.8. Why do models for recombination include the generation of heteroduplexes in the products?
Question 8.9. Consider two DNA duplexes that undergo recombination by the double-strand break mechanism. The parental duplex indicated by thin lines has dominant alleles for genes M, N, O, P, and Q, and the parental duplex shown in thick lines has recessive alleles, indicated by the lower case letters. The recombination intermediate with two Holliday structures is also shown.
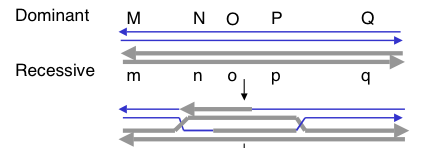
a) What duplexes result from resolution of the left Holliday junction vertically and the right junction horizontally?
b) After the vertical-horizontal resolution, what will the genotype be of the recombination products with respect to the flanking markers M and Q? In answering, use a slash to separate the designation for the 2 chromosomes, each of which is indicated by a line (i.e. the parental arrangement is M___Q / m___q).
c) If the products of the vertical-horizontal resolution were separated by meiosis, and then replicated by mitosis to generate 8 spores in an ordered array (as in the Ascomycete fungi), what would be the phenotype of the spores with respect to alleles of gene O? Assume that the sister chromatids of these chromosomes did not undergo recombination in this region (i.e. one parental duplex from each homologous chromosome remains from the 4n stage).
For the next 3 problems, consider two DNA duplexes that undergo recombination by the double-strand break mechanism. The parental duplex denoted by thin black lines has dominant alleles (capital letters) for genes (or loci) K, L, and M, and the parental duplex denoted by thick gray lines has recessive alleles, indicated by k, l, m. The genes are shown as boxes with gray outlines. In the diagram on the right, the double strand break has been made in the L gene in the black duplex and expanded by the action of exonucleases.
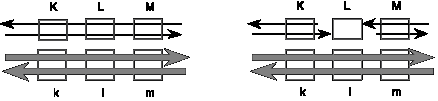
Question 8.10. When recombination proceeds by the double-strand break mechanism, what is the structure of the intermediate with Holliday junctions, prior to branch migration? Please draw the structure, and distinguish between the DNA chains from the parental duplexes.
Question 8.11. If the recombination intermediates are resolved to generate a chromosome with the dominant K allele of the K gene and the recessive m allele of the M gene on the same chromosome (K___m), which allele (dominant L or recessive l) will be be at the L, or middle, gene?
Question 8.12. If the left Holliday junction slid leftward by branch migration all the way through the K gene (K allele on the black duplex, k allele on the gray duplex), what will the structure of the product be, prior to resolution?
Question 8.13. According to the original Holliday model and the double-strand break model for recombination, what are the predicted outcomes of recombination between a linear duplex chromosome and a (formerly) circular duplex carrying a gap in the region of homology? The homology is denoted by the boxes labeled ABC on the linear duplex and ac on the gapped circle. The regions flanking the homology (P and Q versus X and Y) are not homologous.
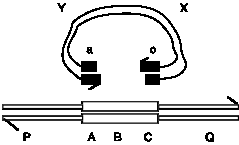
The results of an experiment like this are reported in Orr-Weaver, T. L., Szostak, J. W. and Rothstein, R. J. (1981) Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354-6358. These data were instrumental in formulating the double-strand-break model for recombination.
Question 8.14. A variety of in vitro assays have been developed for strand exchange catalyzed by RecA. For each of the substrates shown below, what are the expected products when incubated with RecA and ATP (and SSB to facilitate removal of secondary structures from single-stranded DNA)? In practice, the reactions proceed in stages and one can see intermediates, but answer in terms of the final products after the reaction has gone to completion.
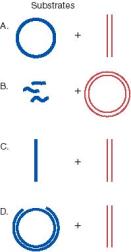
In each case, the molecule with at least partical single stranded region is shown with thick blue strands, and the duplex that will be invaded is shown with thin red lines. The DNA substrates are as follows.
A. Single-stranded circle and duplex linear. The two substrates are the same length and are homologous throughout.
B. Single-stranded short linear fragments and duplex circle. The short fragments are homologous to the circle.
C. Single-stranded linear and duplex linear. The two substrates are the same length and are homologous throughout.
D. Gapped circle and duplex linear. The intact strand of the circle is the same length as the linear and is homologous throughout. The gapped strand of the circle is complementary to the intact strand, of course, but is just shorter.