Description
The Integrative and Discriminative EPigenome Annotation System (IDEAS) method for simultaneous two-dimensional segmentation (Zhang et al. NAR 2016; Zhang and Hardison, NAR 2017) was used to run on 5 epigenetic marks (H3K4me3, H3K4me1, H3K36me3, H3K27me3, H3K9me3) in 127 cell types in the Roadmap Epigenomics project. For this segmentation by IDEAS, the -log10 pval tracks were used as input. We used a pipeline to improve the reproducibility of the functional annotation (Zhang and Hardison, NAR 2017). For this run on 5 features in 127 cell types, the reproducibility cut off was set at 90%.
Display Conventions and Configuration
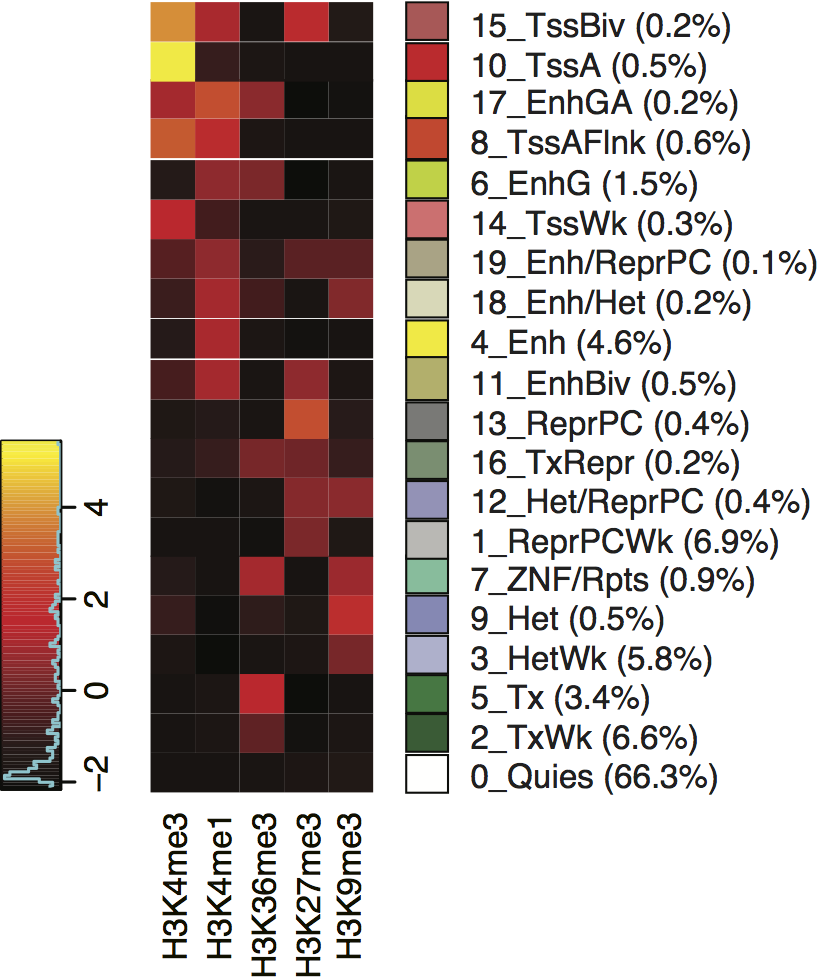
The contributions of each epigenetic feature to each of the 20 states is shown in the figure, along with the color code and their abundance in 127 cell types. Briefly, warm colors are associated with activity and cold colors with inactivity. In more detail, red denotes a state with a promoter-like signature, yellow denote states with an enhancer-like signature, and green denotes a signature for elongating RNA polymerase. In contrast, the grey states convey various combinations of no to low signals for any of the features, and blue states generally correspond to repressive signals.
State Color Key:
Data source
http://egg2.wustl.edu/roadmap/data/byFileType/signal/consolidated/macs2signal/pval/
Contact
Questions should be sent to Yu Zhang at yzz2 at psu dot edu
References
Zhang Y, Ross C Hardison. (2017) Accurate and reproducible functional maps in 127 human cell types via 2D genome segmentation. Nucleic Acids Res. gkx659. DOI:
https://doi.org/10.1093/nar/gkx659
Zhang Y, An L, Yue F, Hardison RC (2016). Jointly characterizing epigenetic dynamics across multiple human cell types. Nucleic Acids Res. 44(14):6721-31.
